Abstract
Reactive oxygen species is one of the most common cellular RNA damaging agents in living organisms. A growing number of studies show a strong correlation between oxidatively damaged RNA and human diseases, predominantly age-related neurodegenerative disorders. Oxidized RNAs impair the fundamental cellular processes including gene regulatory activities and protein synthesis. Molecular characterization of oxidized RNA such as understanding the sources of RNA oxidation, their mechanism of action, and cellular consequences may help to unravel their involvement in the pathogenesis of human diseases. Several proteins and factors with potential function to control RNA oxidation have been identified. Here, we will discuss the role of oxidized RNA binding protein polynucleotide phosphorylase (PNPase) in the quality control of oxidized RNA. PNPase is an evolutionarily conserved 3’-5’ exoribonuclease having multifaceted RNA regulatory roles. Apart from binding to oxidized RNA, PNPase reduces the level of RNA oxidation and protects cells during oxidative stress. In this review, we discuss RNA oxidation and its quality control process with a specific focus on PNPase in regulating oxidized RNA.
Keywords: Reactive oxygen species (ROS), oxidative stress, RNA oxidation, PNPase, quality control pathways, aging, age-related neurodegenerative diseases
Most life forms on earth rely on some forms of oxygen for their existence. Although oxygen is an essential component, it is also metabolized to produce reactive oxygen species (ROS)(1). ROS are highly unstable and reacts with cellular components including the biomolecules in its vicinity. In eukaryotes, ROS are primarily generated as a byproduct of oxidative phosphorylation inside mitochondria. In addition, they are also produced by cellular organelles such as peroxisomes, lysosomes, endoplasmic reticulum (ER)(2,3) and enzymatic reactions in the cell membrane. Exogenous sources of ROS are ionizing and ultraviolet radiations, chemotherapeutic agents, air pollutants, transition metals, and certain foods(3). ROS can be either free radicals such as superoxides (•O2) and hydroxyl radicals (HO•) or nonradical molecules such as hydrogen peroxide (H2O2), and singlet oxygen (1O2). ROS can be beneficial to the cellular system as they trigger the onset of various cell signaling cascades(4,5) and host defense when present at optimum level (6,7). When the levels of ROS exceed the physiologically tolerable range, they can disrupt the fundamental cellular processes such as transcription(8) RNA processing(9), translation(10), and RNA export(11). One of the many reasons of such alteration could be that ROS induces the oxidative modification and damage of cellular biomolecules such as proteins, lipids, DNA and RNA. Such alteration may eventually lead to cell death and trigger the pathogenesis of various human diseases(12,13).
ROS are neutralized by the antioxidant defense mechanism which is also known as the first line of cellular defense against ROS. The defense mechanism involves both enzymatic or non-enzymatic factors(14). Some of the common enzymatic antioxidants are catalase, superoxides dismutase (SOD), and glutathione peroxidases whereas ascorbic acid and lipoic acid, carotenoids, and tocopherols are non-enzymatic antioxidants. Despite the cellular antioxidant defense system, ROS can escape these processes under sub-optimal cellular conditions such as nutrient starvation, endoplasmic reticulum stress, and oxidative stress. Also, ROS can accumulate when the functions of such antioxidants are impaired. Such impairments can lead to disease states. For example, the mutation in superoxide dismutase 1 (SOD1) enhances the level of ROS which is associated with the pathogenesis of Amyotrophic Lateral Sclerosis (ALS) (reviewed in(15)).
Although the oxidative modifications and oxidative damage of most of the biomolecules such as DNA, proteins, and lipids by ROS have been extensively characterized, RNA research has just begun to be recognized. This could be due to the notion that RNAs have a very short half-life which undergo rapid turnover before causing deleterious effects on the cellular system. But some RNAs have a half-life of more than an hour and some even days, hence they have ample opportunities to cause deleterious activities before their turnover. In the subsequent sections, we will discuss how RNA could be targeted by ROS. We then discuss the consequences of such ROS-derived RNA oxidation and highlight its relevance in the pathogenesis of human diseases. Next, we describe the mechanisms by which cells control the level of oxidized RNA, with a specific focus on the role of polynucleotide phosphorylase (PNPase). Finally, we will conclude with open questions that remain to addressed by future studies towards the therapeutic intervention of oxidized RNA associated diseases.
- Reactive oxygen species (ROS) and RNA damage
RNA can be the target of various nucleic acid damaging agents such as ionizing radiation, ultraviolet radiation, xenobiotics, therapeutic drugs, and reactive nitrogen species (RNS), and ROS(16). Damage due to ROS is perhaps the most prominent intracellular source of damage to the RNA(17,18,19). Compared to other biomolecules such as DNA and proteins, RNA is more vulnerable to oxidative damage(20). Such elevated levels of RNA oxidation could be due to that RNA are single-stranded and less protected by binding proteins(19). Also, RNAs are exported in the cytoplasm where the level of ROS is higher than that of the nucleus. In addition, unlike DNA repair mechanisms, no such mechanism has been characterized in repairing oxidatively damaged RNA. ROS can oxidatively modify the bases, phosphate backbone, or sugar moiety of an RNA, however, the modification of bases has been widely characterized. More than 20 different purine and pyrimidine oxidative base modifications have been reported in DNA and this list is growing(21). Since RNA shares the majority of the nucleobases with DNA, it is highly likely that such modifications could also be present in RNA(22). Among many oxidative base modifications, hydroxyl (.OH) induced guanosine base modification, 8-oxoguanosine (8-oxoG) is one of the most commonly characterized and also highly mutagenic base modifications(18). The level of 8-oxoG is also used as a common biomarker of RNA oxidation in most studies. However, the actual level of RNA oxidation should be higher than the level of 8-oxoG as ROS has tendency to oxidatively modify all RNA bases. Moreover, oxidative stress can also cleave the nucleobases that results in the formation of AP RNAs(23). Thus, AP site can be another marker of RNA oxidation which can be measured using aldehyde reactive probe (ARP) assay(24).
Oxidative stress can target all different types of RNA species including transfer RNA (tRNA), ribosomal RNA (rRNA), and messenger RNA (mRNA)(for details see a recent excellent review (25)). The extent of RNA oxidation can vary significantly under oxidative stress and pathological condition(26). In humans, for instance, the level of mRNA and rRNA oxidation were found to be much higher in mild cognitive impairment (MCI), Alzheimer’s Disease (AD), amyotrophic lateral sclerosis (ALS)(27,28,29,30). In E. coli cells, rRNAs are oxidized to a relatively lesser extent than non-ribosomal RNA under normal growth condition(31). Interestingly, when cells were exposed to an exogenous oxidant such as hydrogen peroxide, both the ribosomal and non-ribosomal RNA had an equal increase in the level of RNA oxidation. Moreover, rRNA and non-rRNA in their native and denatured state had an equal increase in the level of RNA oxidation, suggesting that all RNA types can be oxidized regardless of their higher-order structure or their association with proteins. Oxidative stress also can oxidatively damage non-coding RNAs such as microRNA(32). Oxidation of microRNA has been shown to misrecognize its target and leads to apoptosis(33) and cardiac hypertrophy(34). It should be noted that the extent of oxidative modification of microRNAs is not uniform as miR-184(33) and miR-1b34 were preferentially oxidized over other types of miRNAs when they were examined under similar conditions.
- Consequences of RNA oxidation
The robustness and spryness of cellular processes and their proper functioning depend on the quality of their individual components. When the integrity and quality of RNA is threatened by ROS, they alter cellular metabolism and homeostasis. Besides modification of RNA components, oxidative stress also induces RNA strand scission. Exposure of eukaryotic and prokaryotic cells such as E. coli, cultured human cells, and yeast cells to oxidants has been shown to induce cleavage of rRNA(31,35,3637)as well as tRNAs(38,39,40). Increased accumulation of 3’UTR RNA fragments were observed in ageing brains which were associated with ROS(41). Oxidized RNA has also been shown to induce errors in the fundamental cellular processes such as RNA export(11), translation(27,42,43,29,44,45,46), transcription (47, 48, 49, 50), and microRNA mediated gene regulation (32, 33). At the cellular level, a strong correlation between RNA oxidation and cell death has been established (35, 31). Treatment of E. coli or cultured human cells with oxidants such as hydrogen peroxide increases the level of RNA oxidation and decreases cell viability (35, 51). Although the mechanism that links RNA oxidation with cell death has not been well-established, recent studies show that oxidized RNA activates the apoptotic associated pathways. For instance, Ishii and colleagues demonstrated that poly (C)-binding protein 1 (PCBP1) recognizes highly oxidized RNA and activates the apoptosis-related factors such as caspase-3 and cleavage of Poly [ADP-ribose] polymerase 1 (PARP1) (52). Moreover, Tanaka et al showed that cytochrome c (cyt c) cross-links with oxidized RNA and facilitate its dissociation from the mitochondria to the cytoplasm to induce a mitochondrial mediated apoptosis (53). In addition, microRNA mediated oxidation such as miR-184 also triggers the apoptotic pathways by interacting with the 3’UTR of Bcl-xL and Bcl-w (33). In another study in yeast, mutating of decapping enzyme Kluyveromyces lactis Lsm4p (KILsm4P), increased the level of oxidized RNA and induces apoptosis when left unrepaired, they induce apoptosis (54). These findings suggest that oxidized RNA trigger cell death which may be due to their inability to perform many cellular processes including the activation of apoptosis-related pathways.
- RNA Oxidation and human diseases
Oxidized RNAs are strongly associated with a wide range of human diseases, predominantly age-related neurodegenerative disorders (55, 26, 56, 57, 19, 58). The level of RNA oxidation in pathological conditions such as Alzheimer’s disease (AD) (28, 59), Parkinson’s disease (PD) (60), Dementia with Lewy bodies (61), Amyotrophic Lateral Sclerosis (ALS) (29), and multiple sclerosis (62) was found to be significantly higher than that of age-matched healthy controls. These neurodegenerative disorders are commonly associated with neuronal death, which could result from the production of aberrant and truncated proteins due to oxidized mRNAs. In most of these cases, neurons exhibited increased RNA oxidation before their death (29, 63) and such oxidation of RNA is observed in the early stages of these neurodegenerative disorders (64, 29, 63). The brain is one of the metabolically active organs that consumes almost 20% of the total oxygen in the body (65). Higher oxygen metabolism could produce an elevated level of ROS. Moreover, the increased level of ROS could also be associated with a higher number of mitochondria, polyunsaturated fatty acids, modest antioxidant defense system, and a higher level of redox-active transition metals (65, 66). As briefly discussed in the earlier section, the deleterious effect of ROS is controlled by the cellular antioxidant defense system. However, any alternation in the fine balance between ROS level and antioxidant defense mechanism could oxidatively damage the biomolecules such as RNA that ultimately led to these disorders.
(62) was found to be significantly higher than that of age-matched healthy controls. These neurodegenerative disorders are commonly associated with neuronal death, which could result from the production of aberrant and truncated proteins due to oxidized mRNAs. In most of these cases, neurons exhibited increased RNA oxidation before their death (29, 63) and such oxidation of RNA is observed in the early stages of these neurodegenerative disorders (64, 29, 63). The brain is one of the metabolically active organs that consumes almost 20% of the total oxygen in the body (65). Higher oxygen metabolism could produce an elevated level of ROS. Moreover, the increased level of ROS could also be associated with a higher number of mitochondria, polyunsaturated fatty acids, modest antioxidant defense system, and a higher level of redox-active transition metals (65, 66). As briefly discussed in the earlier section, the deleterious effect of ROS is controlled by the cellular antioxidant defense system. However, any alternation in the fine balance between ROS level and antioxidant defense mechanism could oxidatively damage the biomolecules such as RNA that ultimately led to these disorders.
Besides neurodegenerative disorders, increased RNA oxidation level has also been observed in other pathological conditions such as diabetes67, schizophrenia (68, bipolar 1 disorder (BD1) 69, 70) atherosclerosis (71), hereditary hemochromatosis (72), colorectal cancer (73), prion disease (74), Down’s syndrome (75), and aneurysmal subarachnoid haemorrhage (76) indicating that RNA oxidation could be an important event associated with a multitude of human diseases. These findings suggest that RNA oxidation could potentially be used as a diagnostic biomarker for these wide ranges of human diseases (77, 78, 79). Currently, several assays and approaches have been used in the detection and quantification of oxidized RNA. These approaches rely on the use of liquid chromatography coupled with mass spectrometry (LC-MS/MS), comet assay, immunoassay, and ELISA (for details, see the recent review (80)). Multiple human samples such as urine, blood, tissues have been used for the detection and quantification of oxidized RNA.
One of the possible therapeutic approaches of oxidized RNA associated human diseases can be to control the levels of the formation of oxidized RNA in the cellular system. The level of oxidized guanine residue in DNA (8-oxo-dG) was evaluated in response to certain drugs (see review (77)). Plant extract based supplements (81) and antioxidants (82) reduced the levels of DNA/RNA and protein oxidation suggesting that they could help to prevent oxidative processes. Hereafter, we will discuss the cellular mechanism of dealing with oxidatively damaged RNA.
- The quality control mechanism of oxidized RNA
RNA oxidation poses a significant threat to cells and, therefore, cells need to recognize and eliminate them from the cellular system. A number of cellular mechanisms could be involved in the quality control of oxidized RNA (18, 46). Oxidized RNA may be initially recognized and sequestered from the normal RNA pool by a number of oxidized RNA binding proteins and factors (18). In 2001, Hayakawa et.al discovered a high binding affinity of polynucleotide phosphorylase (PNPase) to oxidized RNA while searching for E. coli proteins that bind to oxidized RNA (83). The detailed molecular mechanism of bacterial PNPase and its human homolog (PNPT1) in dealing with oxidized RNA will be discussed in later sections. In 2002, Hayakawa et al further demonstrated yet another protein, Y-box-binding protein (YB-1), having a high binding affinity to RNA containing 8-oxoG (84). Once the oxidized RNAs are recognized and bound by oxidized RNA binding proteins, they may either be repaired by RNA repair enzymes or degraded by ribonucleases [Figure 1]. The final product after the degradation of oxidized RNA may be either re-incorporated during RNA synthesis or eliminated out of the body in the form of urine or other bodily fluids. However, the incorporation of 8-oxoG into new RNA is a rare event as cells have developed several mechanisms to block this process. First, RNA polymerases in both E. coli (85) and human (86) are less efficient in utilizing 8-oxoGTP compared to normal GTP. Second, oxidized nucleoside tri- (8-oxo-GTP) and di - (8-oxo-GDP) phosphates are hydrolyzed to 8-oxo-GMP by several hydrolyzing enzymes such as MutT, MTH1, NUDT5, MTH2, and MTH3 (NUDT18) (reviewed in (18)). Finally, unlike conversion of GMP to GDP by guanylate kinase (GK), it does not convert 8-oxo-GMP to 8-oxo-GDP (86). Aside from decay factors and RNA binding proteins in controlling oxidatively damaged RNA, RNA granules such as stress granules (SG) and P-bodies may have an important role in further processing and eventual decay ( 87). The direct involvement of these RNA granules in the regulation or turnover of oxidatively damaged RNA remains to be characterized.
The cellular mechanisms for handling oxidatively damaged RNA may involve recognition, binding, sequestration, repair, degradation of oxidized RNA, and blocking the incorporation of oxidized ribonucleotides to newly synthesized RNA (18). Recently, two promising discoveries have been made in the quality control of oxidized RNA (46, 88, 89, 90, 91). The first describes a mechanism mediated by the no-go-decay (NGD) pathway (88). During translation, when a ribosome encounters a modified nucleotide such as 8-oxoG, it stalls on that specific position of an mRNA containing such modified nucleobase. This recruits multiple factors that eventually regulate the fate of the nascent truncated polypeptide, the ribosome, and the damaged mRNA. Yan et al showed that modification to even a single nucleotide in the mRNA such as 8-oxoG is enough to activate the NGD pathway (88). Once the NGD pathway is activated, the exoribonuclease XRN1 degrades the mRNA from its 5’-end. Interestingly, they showed that upon deletion of XRN1, there was an increased accumulation of 8-oxoG suggesting an important role of XRN1 in the exonucleolytic digestion of oxidatively damaged RNA. This finding supports the previous reports where disruptions in the NGD pathway led to an increased accumulation of 8-oxoG (42). Recently, it has been discovered that endoribonucleases, CUE2 (92), and NONU-1 (93) are involved in the cleavage of mRNAs near the stalled ribosomes in S. cerevisiae and C. elegans, respectively. These endoribonucleases may also be involved in the cleavage of oxidized mRNAs undergoing ribosome stalling.
The second mechanism involves the RNA binding proteins (RBPs) such as AUF1 and/or PCBP1 [See ref. (18, 94) for the list of all proteins involved in binding and regulating oxidized RNA]. These proteins recognize and bind to oxidized RNA. These RBPs may catalyze oxidized RNA turnover on their own or may recruit other factors. For instance, AUF1 has been shown to detect and bind to moderately oxidized RNA (RNA containing only a few 8-oxoGs) (89) and target them to downstream factors involved in RNA decay pathways (89). On the other hand, PCBP1 recognizes heavily oxidized RNA and induces the activation of factors involved in apoptosis (52, 90). These findings have enhanced our understanding of molecular factors involved in the quality control of oxidized RNA. However additional studies are required to deepen our knowledge of these mechanisms. For instance, the current evidence suggests that no-go decay and ribosome quality control pathways maybe activated when mRNAs are oxidized even at the single nucleotide base modification (88), while AUF1 or PCBP1 may come into play when RNA is moderately and heavily oxidized, respectively (64). More empirical evidence is required to substantiate this observation. Since PNPase is an exoribonuclease, theoretically it may be associated with both the NGD and AUF/PCBP1 pathways in the quality control of oxidized RNA. The byproduct of those pathways may be eventually cleared out from the cellular system with the exoribonucleases as shown in Figure 1. The involvement of exoribonuclease at the 5’ end of RNA pathway has been discussed (88) and PNPase might be another important exoribonuclease at the 3’-end for the cumulative removal of oxidatively damaged RNA. In the subsequent section, we will discuss the multifaceted RNA regulatory role of polynucleotide phosphorylase (PNPase) with a focus on the quality control process of oxidized RNA.
- PNPase in regulating oxidized RNA and cell protection during oxidative stress
PNPase is a 3’-5’ exoribonuclease that belongs to the PDX family of enzymes (95, 96). It was first discovered in Azobacter vinelandii and its function was reported to synthesize polynucleotides (RNA) from 5’-nucleosides diphosphates (dNDPs) (97). Later, the primary function of PNPase was identified to catalyze the phosphorolysis of ribonucleotides from its 3’ to 5’ end (98, 99). PNPase is widely known for its 3’-5’ exoribonuclease activity and its phosphorolysis activity was found to be more favorable in the presence of elevated levels of inorganic phosphates (Pi) (100). However, at the low concentration of inorganic phosphate, PNPase adds a heteropolymeric tail to single-stranded RNA (ssRNA) in a template-independent manner (98). Therefore, PNPase is considered a unique exoribonuclease having a dual function in both the degradation and synthesis of RNA. In E. coli, PNPase forms a degradosome complex that is primarily composed of RNA helicase RhlB, endoribonuclease RNase E, and glycolytic enzyme enolase (101, 102, 103). Such degradosome complex plays an essential role in the degradation and quality control process of structured RNAs. PNPase also works in association with HF-I protein (Hfq) and poly (A) polymerase I (PAP I) as well as independent of its RNA helicase partner RhlB (104). PNPase is an evolutionarily conserved protein in bacteria, plants, flies, mammals, and humans. It consists of five conserved domains, two RNase PH domains (RPH1 and RPH2), two RNA binding domains (KH homology and S1), and an alpha subunit (α) in between RPH1 and RPH2 domains [Figure 2A]. In plants and human, PNPase has additional mitochondrial and chloroplast translocating signals at its C-terminal end. Crystal structure of trimeric hPNPase shows that RPH forms the hexametric ring-like structure whereas KH forms the pore (105). The KH traps the long RNA 3’ tail that is directed to the RNase PH channel for further degradation.
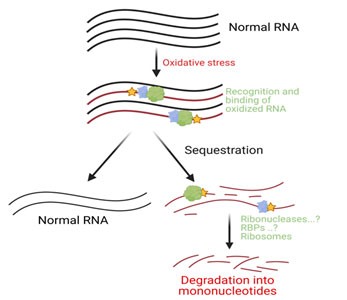
Figure 1. Oxidative stress induces oxidative modifications in RNA. Oxidized RNA may be sequestered from the normal RNA pool by oxidized RNA binding and recognition factors. These sequestered RNAs may eventually undergo turnover due to ribonucleases and ribosomes mediated decay.
Hayakawa and colleagues first demonstrated the involvement of PNPase in the quality control of oxidized RNA in 2001 (83). Using two-dimensional gel analysis in conjunction with the gel-shift assay, they showed that PNPase binds to oligonucleotides containing 8-oxoG with high affinity and protects against ribonuclease RNase A. It is also the first study to show that PNPase responds to cellular oxidative processes as PNPase mutant E. coli strains were hyper resistant to paraquat, a drug that induces oxidative stress. In 2006, the same group demonstrated a similar role of the human homolog of polynucleotide phosphorylase (hPNPase) in binding to 8-oxoG containing RNA (106). These findings provided an important insight into the cellular mechanisms of handling oxidatively damaged RNA. Since PNPase is an exoribonuclease, its high binding affinity to oxidized RNA may eventually degrade oxidized RNAs.
PNPase in cultured human cells reduces the level of 8-oxoG and improves cell viability during oxidative stress (35). Such reduction in 8-oxoG is likely due to degradation of oxidized RNA mediated by hPNPase. Later, the protective role of PNPase against oxidative stress was confirmed in E. coli cell (51). Unlike hyper resistance of PNPase lacking E. coli cells to paraquat (83), Wu and colleagues demonstrated hypersensitivity towards oxidative stress (hydrogen peroxide) (51). Interestingly, such protective function of PNPase is directly associated with the level of RNA oxidation (8-oxoG). A similar role of PNPase has also been characterized in other bacterial strains such as Deinococcus radiodurans (107) and Yersinia pestis although the protective function of the later strain was dependent on its degradosome (108).
As mentioned earlier, hPNPase has an additional mitochondrial translocation signal (MTS) that translocate PNPase to mitochondria (109). Since a majority of the total RNA is present in the cytoplasm, PNPase localization may impact its regulatory role in controlling cytoplasmic oxidized RNA. Also, mitochondria are the predominant intracellular sources of ROS, and they may have a differential effect on RNA inside mitochondria and cytoplasm. For this, the levels of RNA oxidation inside mitochondria and cytoplasm were examined under normal and oxidative stress conditions. The RNA oxidation level was observed to be higher in mitochondrial RNA than that of cytoplasmic RNA as measured by the number of 8-oxoG (110). The knockdown and overexpression of hPNPase showed a significant change in the level of 8-oxoG inside mitochondria and cytoplasm only when cells were exposed to oxidants. This finding further substantiates that PNPase protects cell viability by regulating the level of oxidized RNA during stress conditions. Since hPNPase forms a degradosome complex with human RNA helicase (hSUV3) and the complex is involved in regulating mRNAs inside mitochondria, hSUV3 could also facilitate hPNPase in controlling RNA oxidation (111, 112). However, the knockdown of hSUV3 didn’t change the levels of 8-oxoG under both normal and oxidative stress conditions (110). This suggests that hSUV3 does not have any role in facilitating hPNPase to control oxidized RNA.
PNPase also has evolutionarily conserved domains such as KH and S1 RNA binding domain, RPH1 and RPH2 catalytic domains and alpha subunit [Figure 2A]. These different domains constitute the formation of pores such as S1 pore, KH pore, and RPH pore [Figure 2B]. S1 pore may facilitate the binding of oxidized RNA that may eventually help to direct the oxidized RNA to RPH1 pore for the eventual catalysis and degradation. To provide a further mechanistic understanding of PNPase on oxidatively damaged RNA, the role of an individual domain of PNPase in regulating the 8-oxoG level was examined. Interestingly, the levels of 8-oxoG were significantly increased when cells were transfected with plasmids containing S1, RPH1, and RPH2 deleted hPNPase mutants. The result suggests that the S1 binding domain and RPH1/RPH2 catalytic domains are required for binding, and catalytic activities, respectively (113). Furthermore, a recent computational modeling study that used multi-nanosecond molecular dynamics simulation (114) demonstrated the role of PNPase in recognizing 8-oxoG at the atomic level (115). The study showed that PNPase interacts more favorably with 8-oxoG than non-oxidized Guanosine as per the per-nucleotide energy calculation, stacking contact with specific amino acids of the PNPase subunit, and stronger hydrogen bonding with 2’-OH group. The SFF (S76-F77-F78) groove forms the binding pocket for 8-oxoG that discriminates the 8-oxoG from normal guanosine. Their computationally designed mutational analysis at the SFF groove further reinforces the notion that PNPase has a high binding specificity to 8-oxoG.
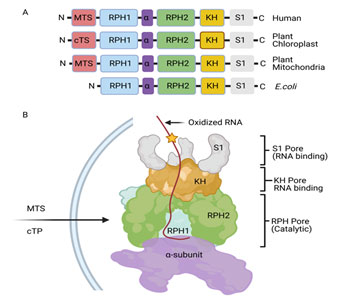
Figure 2. Proposed model of PNPase in controlling RNA oxidation. A. Evolutionarily conserved domains of PNPase in Humans, plants, and E. coli. B. Domain organization of PNPase as described in (116, 117, 105). In humans, MTS will translocate PNPase to mitochondria whereas in plants, MTS and cTP will translocate it to the mitochondria and chloroplast, respectively. The oxidized RNA may be first recognized by S1 and KH RNA binding domains and channel it to RPH catalytic pore for its degradation.
Increased body of evidence supports the notion that PNPase plays a crucial role in controlling oxidized RNA in both prokaryotes and eukaryotes. Biochemical assay (83, 51) and computational simulation at the atomic level (115) show that it preferentially recognizes and binds to 8-oxoG containing RNA with high affinity. Overexpression and knockdown of PNPase in both E. coli and cultured human cells controls the level of 8-oxoG providing firm evidence that it directly regulates oxidized RNA in the cells. Furthermore, cells lacking PNPase were hypersensitive to oxidative stress suggesting that it is an essential ribonuclease in protecting cells under stress conditions. Although it forms a degradosome complex with RNA helicases such as RhlB in E. coli and hSUV3 in eukaryotic cells, these helicases do not seem to facilitate PNPase in controlling oxidized RNA. One of the intriguing features is that PNPase binds to synthetic RNA but does not degrade them 83 whereas it reduces the level of 8-oxoG in vivo (51). Hence, the open question remains whether PNPase recruits other factors to degrade oxidized RNA. PNPase has two RNA binding domains (KH and S1) and two catalytic domains (RPH1 and RPH2). What triggers PNPase’s RNA binding domains to demarcate the binding of oxidized RNA from other aberrant RNA remains to be characterized. PNPase has a dual role as a 3’-5’exoribonuclease and 3’-terminal oligonucleotide polymerases. The reduction of 8-oxoG levels by PNPase inside cells suggests that the exoribonuclease activity is dominant over polymerase activity. However, PNPase may recruit other factors to degrade oxidized RNA after its binding. Hence, the role of other proteins cannot be undermined unless direct evidence of PNPase exoribonuclease activity on oxidized RNA is established.
- Conclusion and future perspectives
An increasing number of studies show that RNA oxidation poses a significant threat to living organisms. RNA oxidation could be a major player of human aging (58) and various diseases. This could be due to the defects in molecular and cellular processes associated with oxidized RNAs such as protein translation, RNA strand scission, abasic site formation, and cell death. Yet, the relationship between RNA oxidation-associated disease and their clinical applications is not fully established. A better understanding of the role of oxidized RNA in human diseases could be discerned by studying the distribution pattern of oxidized RNA (8-oxoG) as has been done for DNA. In DNA, the profiling of oxidized deoxyguanosine (8-oxodG) modification was first studied by Nakabeppu and colleagues in 2006 (118). Subsequent studies show that the sites of 8-oxodG modifications are not consistent (119) instead they are enriched in the gene body and promoter regions when compared to the intergenic regions (5’UTR and 3’UTR) (120, 121, 122, 123).
Similar information for RNA could enhance our understanding of the role of RNA oxidation in various diseases. In RNA, this type of global analysis of the distribution of oxidation has been reported by few studies in yeast (124) and lung cells (125). Further studies using approaches such as high throughput long-read RNA sequencing strategies (third-generation sequencing) could significantly enhance our understanding of RNA oxidation and human diseases
As for the proteins and factors involved in the quality control mechanisms of oxidized RNA, a handful has been well characterized while others are still being discovered. Current evidence points to a crucial role of PNPase. PNPase. Studies in both E. coli and cultured human cells have shown that PNPase is involved in controlling oxidized RNA and thus resulting in cell protection during oxidative stress. Functional defects of PNPase, for instance, due to mutation may interfere with its role in controlling oxidized. This may result in the increased accumulation of oxidized RNA which may lead to, or further, aggravate the pathogenesis of oxidized RNA-associated human diseases.
The growing evidence of the association of RNA oxidation with multiple human diseases warrants further studies into this area. Future studies can probe into how oxidized RNA can be employed as a routine clinical biomarker of various diseases. Also, since PNPase plays an important role in the quality control of oxidized RNAs, further studies should be directed at fully deciphering the pathways associated with PNPase. This can also inform the development of new therapeutic agents that enhance the function of PNPase.
Acknowledgments: We thank the members of Emmanouil Maragkakis lab for helpful discussion on this manuscript.
Abbreviations/nomenclature
1O2 Singlet oxygen
8-oxoG 8-hydroxyguanosine
AD Alzheimer's disease
ALS Amyotrophic lateral sclerosis
ARP Aldehyde reactive probe assay
ATP Adenosine triphosphate
AUF1 AU-rich element RNA-binding protein 1
BD1 Bipolar disorder 1
CNS Central nervous system
DAZAP1 DAZ Associated Protein 1
DNA Deoxyribonucleic acid
E. coli Escherichia coli
ER Endoplasmic reticulum
ES7 Expansion segment 7
H2O2 Hydrogen peroxide
HO• Hydroxyl groups
HPLC High-performance liquid chromatography
hSUV3 Human RNA helicase
Kilsm4p Kluyveromyces lactis Lsm4p
Lsm4p U6 snRNA-associated Sm-like protein
mRNA Messenger RNA
MTS Mitochondrial translocation signal
O2 _ Superoxide
ONOO− Peroxynitrite
PARP1 Poly [ADP-ribose] polymerase 1
PCBP1 poly C binding protein one
PD Parkinson's disease
PNPase polynucleotide phosphorylase
RNA Ribonucleic acid
RNS Reactive nitrogen species
ROS Reactive oxygen species
rRNA Ribosomal RNA
ssRNA Single stranded RNA
tRNA Transfer RNA
UTR Untranslated region
UV Ultraviolet
References:
|
1. Markkanen, E. Not breathing is not an option: How to deal with oxidative DNA damage. DNA Repair (Amst.) 59, 82-105 (2017).
https://doi.org/10.1016/j.dnarep.2017.09.007 PMid:28963982
|
|
2. Murphy, M. P. How mitochondria produce reactive oxygen species. Biochem. J. 417, 1-13 (2009). https://doi.org/10.1042/BJ20081386 PMid:19061483 PMCid:PMC2605959
|
|
|
3. Di Meo, S., Reed, T. T., Venditti, P. & Victor, V. M. Role of ROS and RNS sources in physiological and pathological conditions. Oxid. Med. Cell. Longev. 2016, 1245049 (2016). https://doi.org/10.1155/2016/1245049 PMid:27478531 PMCid:PMC4960346
|
|
|
4. Rhee, S. G. Cell signaling. H2O2, a necessary evil for cell signaling. Science 312, 1882-1883 (2006). https://doi.org/10.1126/science.1130481 PMid:16809515
|
|
|
5. Lanz, M. C., Dibitetto, D. & Smolka, M. B. DNA damage kinase signaling: checkpoint and repair at 30 years. EMBO J. 38, e101801 (2019).
https://doi.org/10.15252/embj.2019101801 PMid:31393028 PMCid:PMC6745504
|
|
|
6. Silwal, P., Kim, J. K., Kim, Y. J. & Jo, E.-K. Mitochondrial reactive oxygen species: Double-edged weapon in host defense and pathological inflammation during infection. Front. Immunol. 11, 1649 (2020). https://doi.org/10.3389/fimmu.2020.01649 PMid:32922385 PMCid:PMC7457135
|
|
|
7. Paiva, C. N. & Bozza, M. T. Are reactive oxygen species always detrimental to pathogens? Antioxid. Redox Signal. 20, 1000-1037 (2014).
https://doi.org/10.1089/ars.2013.5447 PMid:23992156 PMCid:PMC3924804
|
|
|
8. Ma, Q. Transcriptional responses to oxidative stress: pathological and toxicological implications. Pharmacol. Ther. 125, 376-393 (2010).
https://doi.org/10.1016/j.pharmthera.2009.11.004PMid:19945483
|
|
|
9. Szaflarski, W. et al. A novel stress response pathway regulates rRNA biogenesis. bioRxiv (2020) doi:10.1101/2020.08.16.250183. https://doi.org/10.1101/2020.08.16.250183
|
|
|
10. Ghosh, A. & Shcherbik, N. Effects of oxidative stress on protein translation: Implications for cardiovascular diseases. Int. J. Mol. Sci. 21, 2661 (2020).
https://doi.org/10.3390/ijms21082661 PMid:32290431 PMCid:PMC7215667
|
|
|
11. Schwenzer, H. et al. Oxidative Stress Triggers Selective tRNA Retrograde Transport in Human Cells during the Integrated Stress Response. Cell Rep. 26, 3416-3428.e5 (2019). https://doi.org/10.1016/j.celrep.2019.02.077 PMid:30893612 PMCid:PMC6426654
|
|
|
12. Liu, Z., Zhou, T., Ziegler, A. C., Dimitrion, P. & Zuo, L. Oxidative stress in neurodegenerative diseases: From molecular mechanisms to clinical applications. Oxid. Med. Cell. Longev. 2017, 2525967 (2017). https://doi.org/10.1155/2017/2525967 PMid:28785371 PMCid:PMC5529664
|
|
|
13. Martindale, J. L. & Holbrook, N. J. Cellular response to oxidative stress: signaling for suicide and survival. J. Cell. Physiol. 192, 1-15 (2002). https://doi.org/10.1002/jcp.10119 PMid:12115731
|
|
|
14. Birben, E., Sahiner, U. M., Sackesen, C., Erzurum, S. & Kalayci, O. Oxidative stress and antioxidant defense. World Allergy Organ. J. 5, 9-19 (2012).
https://doi.org/10.1097/WOX.0b013e3182439613 PMid:23268465 PMCid:PMC3488923
|
|
|
15. Kaur, S. J., McKeown, S. R. & Rashid, S. Mutant SOD1 mediated pathogenesis of Amyotrophic Lateral Sclerosis. Gene 577, 109-118 (2016).
https://doi.org/10.1016/j.gene.2015.11.049 PMid:26657039
|
|
|
16. Wurtmann, E. J. & Wolin, S. L. RNA under attack: cellular handling of RNA damage. Crit. Rev. Biochem. Mol. Biol. 44, 34-49 (2009).
https://doi.org/10.1080/10409230802594043 PMid:19089684 PMCid:PMC2656420
|
|
|
17. Li, Z., Wu, J. & Deleo, C. J. RNA damage and surveillance under oxidative stress. IUBMB Life 58, 581-588 (2006). https://doi.org/10.1080/15216540600946456 PMid:17050375
|
|
|
18. Li, Z., Malla, S., Shin, B. & Li, J. M. Battle against RNA oxidation: molecular mechanisms for reducing oxidized RNA to protect cells. Wiley Interdiscip. Rev. RNA 5, 335-346 (2014). https://doi.org/10.1002/wrna.1214 PMid:24375979 PMCid:PMC3991771
|
|
|
19. Fimognari, C. Role of oxidative RNA damage in chronic-degenerative diseases. Oxid. Med. Cell. Longev. 2015, 358713 (2015). https://doi.org/10.1155/2015/358713 PMid:26078805 PMCid:PMC4452857
|
|
|
20. Hofer, T. et al. Hydrogen peroxide causes greater oxidation in cellular RNA than in DNA. Biol. Chem. 386, 333-337 (2005). https://doi.org/10.1515/BC.2005.040 PMid:15899695
|
|
|
21. Barciszewski, J., Barciszewska, M. Z., Siboska, G., Rattan, S. I. & Clark, B. F. Some unusual nucleic acid bases are products of hydroxyl radical oxidation of DNA and RNA. Mol. Biol. Rep. 26, 231-238 (1999). https://doi.org/10.1023/A:1007058602594 PMid:10634505
|
|
|
22. Gajewski, E., Rao, G., Nackerdien, Z. & Dizdaroglu, M. Modification of DNA bases in mammalian chromatin by radiation-generated free radicals. Biochemistry 29, 7876-7882 (1990). https://doi.org/10.1021/bi00486a014 PMid:2261442
|
|
|
23. Tanaka, M., Han, S., Küpfer, P. A., Leumann, C. J. & Sonntag, W. E. Quantification of oxidized levels of specific RNA species using an aldehyde reactive probe. Anal. Biochem. 417, 142-148 (2011). https://doi.org/10.1016/j.ab.2011.05.038 PMid:21693097 PMCid:PMC3143300
|
|
|
24. Tanaka, M., Han, S., Küpfer, P. A., Leumann, C. J. & Sonntag, W. E. An assay for RNA oxidation induced abasic sites using the Aldehyde Reactive Probe. Free Radic. Res. 45, 237-247 (2011). https://doi.org/10.3109/10715762.2010.535529 PMid:21062214 PMCid:PMC3058411
|
|
|
25. Tanaka, M. & Chock, P. B. Oxidative modifications of RNA and its potential roles in biosystem. Front. Mol. Biosci. 8, 685331 (2021).
https://doi.org/10.3389/fmolb.2021.685331 PMid:34055897 PMCid:PMC8149912
|
|
|
26. Kong, Q. & Lin, C.-L. G. Oxidative damage to RNA: mechanisms, consequences, and diseases. Cell. Mol. Life Sci. 67, 1817-1829 (2010).
https://doi.org/10.1007/s00018-010-0277-y PMid:20148281 PMCid:PMC3010397
|
|
|
27. Shan, X., Tashiro, H. & Lin, C.-L. G. The identification and characterization of oxidized RNAs in Alzheimer's disease. J. Neurosci. 23, 4913-4921 (2003).
https://doi.org/10.1523/JNEUROSCI.23-12-04913.2003 PMid:12832513 PMCid:PMC6741200
|
|
|
28. Shan, X. & Lin, C.-L. G. Quantification of oxidized RNAs in Alzheimer's disease. Neurobiol. Aging 27, 657-662 (2006). https://doi.org/10.1016/j.neurobiolaging.2005.03.022 PMid:15979765
|
|
|
29. Chang, Y. et al. Messenger RNA oxidation occurs early in disease pathogenesis and promotes motor neuron degeneration in ALS. PLoS One 3, e2849 (2008). https://doi.org/10.1371/journal.pone.0002849 PMid:18682740 PMCid:PMC2481395
|
|
|
30. Ding, Q., Markesbery, W. R., Chen, Q., Li, F. & Keller, J. N. Ribosome dysfunction is an early event in Alzheimer's disease. J. Neurosci. 25, 9171-9175 (2005). https://doi.org/10.1523/JNEUROSCI.3040-05.2005 PMid:16207876 PMCid:PMC6725754
|
|
|
31. Liu, M. et al. Characterization of RNA damage under oxidative stress in Escherichia coli. Biol. Chem. 393, 123-132 (2012). https://doi.org/10.1515/hsz-2011-0247 PMid:22718628 PMCid:PMC3404489
|
|
|
32. Nunomura, A. & Perry, G. RNA and oxidative stress in Alzheimer's disease: Focus on microRNAs. Oxid. Med. Cell. Longev. 2020, 2638130 (2020).
https://doi.org/10.1155/2020/2638130 PMid:33312335 PMCid:PMC7721489
|
|
|
33. Wang, J.-X. et al. Oxidative modification of miR-184 enables it to target Bcl-xL and Bcl-w. Mol. Cell 59, 50-61 (2015). https://doi.org/10.1016/j.molcel.2015.05.003 PMid:26028536
|
|
|
34. Seok, H. et al. Position-specific oxidation of miR-1 encodes cardiac hypertrophy. Nature 584, 279-285 (2020). https://doi.org/10.1038/s41586-020-2586-0
PMid:32760005
|
|
|
35. Wu, J. & Li, Z. Human polynucleotide phosphorylase reduces oxidative RNA damage and protects HeLa cell against oxidative stress. Biochem. Biophys. Res. Commun. 372, 288-292 (2008). https://doi.org/10.1016/j.bbrc.2008.05.058 PMid:18501193 PMCid:PMC2531134
|
|
|
36. Shedlovskiy, D., Zinskie, J. A., Gardner, E., Pestov, D. G. & Shcherbik, N. Endonucleolytic cleavage in the expansion segment 7 of 25S rRNA is an early marker of low-level oxidative stress in yeast. J. Biol. Chem. 292, 18469-18485 (2017). https://doi.org/10.1074/jbc.M117.800003 PMid:28939771 PMCid:PMC5682959
|
|
|
37. Zinskie, J. A. et al. Iron-dependent cleavage of ribosomal RNA during oxidative stress in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 293, 14237-14248 (2018). https://doi.org/10.1074/jbc.RA118.004174 PMid:30021840 PMCid:PMC6139556
|
|
|
38. Thompson, D. M., Lu, C., Green, P. J. & Parker, R. tRNA cleavage is a conserved response to oxidative stress in eukaryotes. RNA 14, 2095-2103 (2008).
https://doi.org/10.1261/rna.1232808 PMid:18719243 PMCid:PMC2553748
|
|
|
39. Fu, H. et al. Stress induces tRNA cleavage by angiogenin in mammalian cells. FEBS Lett. 583, 437-442 (2009). https://doi.org/10.1016/j.febslet.2008.12.043
PMid:19114040
|
|
|
40. Yamasaki, S., Ivanov, P., Hu, G.-F. & Anderson, P. Angiogenin cleaves tRNA and promotes stress-induced translational repression. J. Cell Biol. 185, 35-42 (2009). https://doi.org/10.1083/jcb.200811106 PMid:19332886 PMCid:PMC2700517
|
|
| |
|
41. Sudmant, P. H., Lee, H., Dominguez, D., Heiman, M. & Burge, C. B. Widespread Accumulation of Ribosome-Associated Isolated 3' UTRs in Neuronal Cell Populations of the Aging Brain. Cell Rep. 25, 2447-2456.e4 (2018). https://doi.org/10.1016/j.celrep.2018.10.094 PMid:30485811 PMCid:PMC6354779
|
|
|
42. Simms, C. L., Hudson, B. H., Mosior, J. W., Rangwala, A. S. & Zaher, H. S. An active role for the ribosome in determining the fate of oxidized mRNA. Cell Rep. 9, 1256-1264 (2014). https://doi.org/10.1016/j.celrep.2014.10.042 PMid:25456128 PMCid:PMC4254665
|
|
|
43. Tanaka, M., Chock, P. B. & Stadtman, E. R. Oxidized messenger RNA induces translation errors. Proc. Natl. Acad. Sci. U. S. A. 104, 66-71 (2007).
https://doi.org/10.1073/pnas.0609737104 PMid:17190801 PMCid:PMC1765478
|
|
|
44. Thomas, E. N., Simms, C. L., Keedy, H. E. & Zaher, H. S. Insights into the base-pairing preferences of 8-oxoguanosine on the ribosome. Nucleic Acids Res. 47, 9857-9870 (2019). https://doi.org/10.1093/nar/gkz701 PMid:31400119 PMCid:PMC6765139
|
|
|
45. Dai, D.-P. et al. Transcriptional mutagenesis mediated by 8-oxoG induces translational errors in mammalian cells. Proc. Natl. Acad. Sci. U. S. A. 115, 4218-4222 (2018). https://doi.org/10.1073/pnas.1718363115 PMid:29610342 PMCid:PMC5910836
|
|
|
46. Yan, L. L. & Zaher, H. S. How do cells cope with RNA damage and its consequences? J. Biol. Chem. 294, 15158-15171 (2019).
https://doi.org/10.1074/jbc.REV119.006513 PMid:31439666 PMCid:PMC6791314
|
|
|
47. Rhee, Y., Valentine, M. R. & Termini, J. Oxidative base damage in RNA detected by reverse transcriptase. Nucleic Acids Res. 23, 3275-3282 (1995).
https://doi.org/10.1093/nar/23.16.3275 PMid:7545285 PMCid:PMC307188
|
|
|
48. Gong, X., Tao, R. & Li, Z. Quantification of RNA damage by reverse transcription polymerase chain reactions. Anal. Biochem. 357, 58-67 (2006).
https://doi.org/10.1016/j.ab.2006.06.025 PMid:16860776
|
|
|
49. Alenko, A., Fleming, A. M. & Burrows, C. J. Reverse transcription past products of guanine oxidation in RNA leads to insertion of A and C opposite 8-oxo-7,8-dihydroguanine and A and G opposite 5-guanidinohydantoin and spiroiminodihydantoin diastereomers. Biochemistry 56, 5053-5064 (2017).
https://doi.org/10.1021/acs.biochem.7b00730 PMid:28845978 PMCid:PMC5623583
|
|
|
50. Glennon, M. M., Skinner, A., Krutsinger, M. & Resendiz, M. J. E. Translesion synthesis by AMV, HIV, and MMLVreverse transcriptases using RNA templates containing inosine, guanosine, and their 8-oxo-7,8-dihydropurine derivatives. PLoS One 15, e0235102 (2020). https://doi.org/10.1371/journal.pone.0235102
PMid:32857764 PMCid:PMC7455023
|
|
|
51. Wu, J. et al. Polynucleotide phosphorylase protects Escherichia coli against oxidative stress. Biochemistry 48, 2012-2020 (2009).
https://doi.org/10.1021/bi801752p PMid:19219992 PMCid:PMC2697445
|
|
|
52. Ishii, T., Hayakawa, H., Igawa, T., Sekiguchi, T. & Sekiguchi, M. Specific binding of PCBP1 to heavily oxidized RNA to induce cell death. Proc. Natl. Acad. Sci. U. S. A. 115, 6715-6720 (2018). https://doi.org/10.1073/pnas.1806912115 PMid:29891675 PMCid:PMC6042155
|
|
|
53. Tanaka, M. et al. RNA oxidation catalyzed by cytochrome c leads to its depurination and cross-linking, which may facilitate cytochrome c release from mitochondria. Free Radic. Biol. Med. 53, 854-862 (2012). https://doi.org/10.1016/j.freeradbiomed.2012.05.044 PMid:22683603 PMCid:PMC4319184
|
|
|
54. Stirpe, M. et al. Increased levels of RNA oxidation enhance the reversion frequency in aging pro-apoptotic yeast mutants. Apoptosis 22, 200-206 (2017).
https://doi.org/10.1007/s10495-016-1319-1 PMid:27803986 PMCid:PMC5306349
|
|
|
55. Kong, Q., Shan, X., Chang, Y., Tashiro, H. & Lin, C.-L. G. RNA oxidation: a contributing factor or an epiphenomenon in the process of neurodegeneration. Free Radic. Res. 42, 773-777 (2008). https://doi.org/10.1080/10715760802311187 PMid:18661427
|
|
|
56. Poulsen, H. E. et al. RNA modifications by oxidation: a novel disease mechanism? Free Radic. Biol. Med. 52, 1353-1361 (2012).
https://doi.org/10.1016/j.freeradbiomed.2012.01.009 PMid:22306201
|
|
|
57. Nunomura A. Role of oxidative RNA damage in aging and neurodegenerative disorders. Brain Nerve 65, 179-194 (2013).
|
|
|
58. Luo, J., Mills, K., le Cessie, S., Noordam, R. & van Heemst, D. Ageing, age-related diseases and oxidative stress: What to do next? Ageing Res. Rev. 57, 100982 (2020). https://doi.org/10.1016/j.arr.2019.100982 PMid:31733333
|
|
|
59. Nunomura, A. et al. RNA oxidation is a prominent feature of vulnerable neurons in Alzheimer's disease. J. Neurosci. 19, 1959-1964 (1999).
https://doi.org/10.1523/JNEUROSCI.19-06-01959.1999 PMid:10066249 PMCid:PMC6782583
|
|
|
60. Zhang, J. et al. Parkinson's disease is associated with oxidative damage to cytoplasmic DNA and RNA in substantia nigra neurons. Am. J. Pathol. 154, 1423-1429 (1999). https://doi.org/10.1016/S0002-9440(10)65396-5
|
|
|
61. Nunomura, A. et al. Neuronal RNA oxidation is a prominent feature of dementia with Lewy bodies. Neuroreport 13, 2035-2039 (2002).
https://doi.org/10.1097/00001756-200211150-00009 PMid:12438921
|
|
|
62. Kharel, P., McDonough, J. & Basu, S. Evidence of extensive RNA oxidation in normal appearing cortex of multiple sclerosis brain. Neurochem. Int. 92, 43-48 (2016). https://doi.org/10.1016/j.neuint.2015.12.002 PMid:26706235
|
|
|
63. Shan, X., Chang, Y. & Lin, C.-L. G. Messenger RNA oxidation is an early event preceding cell death and causes reduced protein expression. FASEB J. 21, 2753-2764 (2007). https://doi.org/10.1096/fj.07-8200com PMid:17496160
|
|
|
64. Ding, Q., Markesbery, W. R., Cecarini, V. & Keller, J. N. Decreased RNA, and increased RNA oxidation, in ribosomes from early Alzheimer's disease. Neurochem. Res. 31, 705-710 (2006). https://doi.org/10.1007/s11064-006-9071-5 PMid:16770743
|
|
|
65. Cobley, J. N., Fiorello, M. L. & Bailey, D. M. 13 reasons why the brain is susceptible to oxidative stress. Redox Biol 15, 490-503 (2018).
https://doi.org/10.1016/j.redox.2018.01.008 PMid:29413961 PMCid:PMC5881419
|
|
|
66. Wang, X. & Michaelis, E. K. Selective neuronal vulnerability to oxidative stress in the brain. Front. Aging Neurosci. 2, 12 (2010).
https://doi.org/10.3389/fnagi.2010.00012 PMid:20552050 PMCid:PMC2874397
|
|
|
67. Cejvanovic, V. et al. RNA oxidation and iron levels in patients with diabetes. Free Radic. Biol. Med. 129, 532-536 (2018).
https://doi.org/10.1016/j.freeradbiomed.2018.10.420 PMid:30339885
|
|
|
68. Jorgensen, A. et al. Systemic oxidatively generated DNA/RNA damage in clinical depression: associations to symptom severity and response to electroconvulsive therapy. J. Affect. Disord. 149, 355-362 (2013). https://doi.org/10.1016/j.jad.2013.02.011 PMid:23497793
|
|
|
69. Munkholm, K., Poulsen, H. E., Kessing, L. V. & Vinberg, M. Elevated levels of urinary markers of oxidatively generated DNA and RNA damage in bipolar disorder. Bipolar Disord. 17, 257-268 (2015). https://doi.org/10.1111/bdi.12245 PMid:25118140
|
|
|
70. Jacoby, A. S., Vinberg, M., Poulsen, H. E., Kessing, L. V. & Munkholm, K. Increased DNA and RNA damage by oxidation in patients with bipolar I disorder. Transl. Psychiatry 6, e867-e867 (2016). https://doi.org/10.1038/tp.2016.141 PMid:27505230 PMCid:PMC5022087
|
|
|
71. Martinet, W., de Meyer, G. R. Y., Herman, A. G. & Kockx, M. M. Reactive oxygen species induce RNA damage in human atherosclerosis. Eur. J. Clin. Invest. 34, 323-327 (2004). https://doi.org/10.1111/j.1365-2362.2004.01343.x PMid:15147328
|
|
|
72. Broedbaek, K. et al. Urinary excretion of biomarkers of oxidatively damaged DNA and RNA in hereditary hemochromatosis. Free Radic. Biol. Med. 47, 1230-1233 (2009). https://doi.org/10.1016/j.freeradbiomed.2009.08.004 PMid:19686840
|
|
|
73. Guo, C. et al. 8-Hydroxyguanosine as a possible RNA oxidative modification marker in urine from colorectal cancer patients: Evaluation by ultra performance liquid chromatography-tandem mass spectrometry. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 1136, 121931 (2020).
https://doi.org/10.1016/j.jchromb.2019.121931 PMid:31855840
|
|
|
74. Guentchev, M. et al. Oxidative damage to nucleic acids in human prion disease. Neurobiol. Dis. 9, 275-281 (2002). https://doi.org/10.1006/nbdi.2002.0477
PMid:11950273
|
|
|
75. Nunomura, A. et al. Neuronal RNA oxidation in Alzheimer's disease and Down's syndrome. Ann. N. Y. Acad. Sci. 893, 362-364 (1999).
https://doi.org/10.1111/j.1749-6632.1999.tb07855.x PMid:10672267
|
|
|
76. Jorgensen, A. et al. Progressive DNA and RNA damage from oxidation after aneurysmal subarachnoid haemorrhage in humans. Free Radic. Res. 52, 51-56 (2018). https://doi.org/10.1080/10715762.2017.1407413 PMid:29157018
|
|
|
77. Guo, C. et al. Potential application of the oxidative nucleic acid damage biomarkers in detection of diseases. Oncotarget 8, 75767-75777 (2017).
https://doi.org/10.18632/oncotarget.20801 PMid:29088908 PMCid:PMC5650463
|
|
|
78. Jacob, K. D., Noren Hooten, N., Trzeciak, A. R. & Evans, M. K. Markers of oxidant stress that are clinically relevant in aging and age-related disease. Mech. Ageing Dev. 134, 139-157 (2013). https://doi.org/10.1016/j.mad.2013.02.008 PMid:23428415 PMCid:PMC3664937
|
|
|
79. Larsen, E. L., Weimann, A. & Poulsen, H. E. Interventions targeted at oxidatively generated modifications of nucleic acids focused on urine and plasma markers. Free Radic. Biol. Med. 145, 256-283 (2019). https://doi.org/10.1016/j.freeradbiomed.2019.09.030 PMid:31563634
|
|
|
80. Chao, M.-R. et al. Biomarkers of nucleic acid oxidation - A summary state-of-the-art. Redox Biol. 42, 101872 (2021). https://doi.org/10.1016/j.redox.2021.101872 PMid:33579665 PMCid:PMC8113048
|
|
|
81. Fragopoulou, E. et al. Suppression of DNA/RNA and protein oxidation by dietary supplement which contains plant extracts and vitamins: a randomized, double-blind, placebo-controlled trial. Lipids Health Dis. 17, 187 (2018). https://doi.org/10.1186/s12944-018-0836-z PMid:30115068 PMCid:PMC6097198
|
|
|
82. Gutierrez-Mariscal, F. M. et al. Mediterranean diet supplemented with coenzyme Q10 induces postprandial changes in p53 in response to oxidative DNA damage in elderly subjects. Age (Dordr.) 34, 389-403 (2012). https://doi.org/10.1007/s11357-011-9229-1 PMid:21404051 PMCid:PMC3312623
|
|
|
83. Hayakawa, H., Kuwano, M. & Sekiguchi, M. Specific binding of 8-oxoguanine-containing RNA to polynucleotide phosphorylase protein. Biochemistry 40, 9977-9982 (2001). https://doi.org/10.1021/bi010595q PMid:11502194
|
|
|
84. Hayakawa, H. et al. Binding capacity of human YB-1 protein for RNA containing 8-oxoguanine. Biochemistry 41, 12739-12744 (2002). https://doi.org/10.1021/bi0201872 PMid:12379116
|
|
|
85. Taddei, F. et al. Counteraction by MutT protein of transcriptional errors caused by oxidative damage. Science 278, 128-130 (1997).
https://doi.org/10.1126/science.278.5335.128 PMid:9311918
|
|
|
86. Hayakawa, H. et al. Metabolic fate of oxidized guanine ribonucleotides in mammalian cells. Biochemistry 38, 3610-3614 (1999). https://doi.org/10.1021/bi982361l PMid:10090747
|
|
|
87. Alluri, R. K., Li, Z. & McCrae, K. Stress granule mediated oxidized RNA decay in P-body: Hypothetical role of ADAR1, Tudor-SN and STAU1. Front. Mol. Biosci. 8, 480 (2021). https://doi.org/10.3389/fmolb.2021.672988 PMid:34150849 PMCid:PMC8211916
|
|
|
88. Yan, L. L., Simms, C. L., McLoughlin, F., Vierstra, R. D. & Zaher, H. S. Oxidation and alkylation stresses activate ribosome-quality control. Nat. Commun. 10, 5611 (2019). https://doi.org/10.1038/s41467-019-13579-3 PMid:31819057 PMCid:PMC6901537
|
|
|
89. Ishii, T., Hayakawa, H., Sekiguchi, T., Adachi, N. & Sekiguchi, M. Role of Auf1 in elimination of oxidatively damaged messenger RNA in human cells. Free Radic. Biol. Med. 79, 109-116 (2015). https://doi.org/10.1016/j.freeradbiomed.2014.11.018 PMid:25486179
|
|
|
90. Ishii, T. & Sekiguchi, M. Two ways of escaping from oxidative RNA damage: Selective degradation and cell death. DNA Repair (Amst.) 81, 102666 (2019).
https://doi.org/10.1016/j.dnarep.2019.102666 PMid:31326364
|
|
|
91. Ishii, T. et al. PCBP1 and PCBP2 both bind heavily oxidized RNA but cause opposing outcomes, suppressing or increasing apoptosis under oxidative conditions. J. Biol. Chem. 295, 12247-12261 (2020). https://doi.org/10.1074/jbc.RA119.011870 PMid:32647012
|
|
|
92. D'Orazio, K. N. et al. The endonuclease Cue2 cleaves mRNAs at stalled ribosomes during No Go Decay. Elife 8, e49117 (2019). https://doi.org/10.7554/eLife.49117 PMid:31219035 PMCid:PMC6598757
|
|
|
93. Glover, M. L. et al. NONU-1 encodes a conserved endonuclease required for mRNA translation surveillance. Cell Rep. 30, 4321-4331.e4 (2020).
https://doi.org/10.1016/j.celrep.2020.03.023 PMid:32234470 PMCid:PMC7184879
|
|
|
94. Hayakawa, H. et al. Human proteins that specifically bind to 8-oxoguanine-containing RNA and their responses to oxidative stress. Biochem. Biophys. Res. Commun. 403, 220-224 (2010). https://doi.org/10.1016/j.bbrc.2010.11.011 PMid:21073862
|
|
|
95. Zuo, Y. & Deutscher, M. P. Exoribonuclease superfamilies: structural analysis and phylogenetic distribution. Nucleic Acids Res. 29, 1017-1026 (2001).
https://doi.org/10.1093/nar/29.5.1017 PMid:11222749 PMCid:PMC56904
|
|
|
96. Dos Santos, R. F. et al. Major 3'-5' exoribonucleases in the metabolism of coding and non-coding RNA. Prog. Mol. Biol. Transl. Sci. 159, 101-155 (2018).
https://doi.org/10.1016/bs.pmbts.2018.07.005 PMid:30340785
|
|
|
97. Grunberg-Manago, M., Ortiz, P. J. & Ochoa, S. Enzymic synthesis of polynucleotides. I. Polynucleotide phosphorylase of azotobacter vinelandii. Biochim. Biophys. Acta 20, 269-285 (1956). https://doi.org/10.1016/0006-3002(56)90286-4
|
|
|
98. Mohanty, B. K. & Kushner, S. R. Polynucleotide phosphorylase functions both as a 3' right-arrow 5' exonuclease and a poly(A) polymerase in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 97, 11966-11971 (2000). https://doi.org/10.1073/pnas.220295997 PMid:11035800 PMCid:PMC17278
|
|
|
99. Yehudai-Resheff, S., Hirsh, M. & Schuster, G. Polynucleotide phosphorylase functions as both an exonuclease and a poly(A) polymerase in spinach chloroplasts. Mol. Cell. Biol. 21, 5408-5416 (2001). https://doi.org/10.1128/MCB.21.16.5408-5416.2001 PMid:11463823 PMCid:PMC87263
|
|
|
100. Deutscher, M. P. Degradation of RNA in bacteria: comparison of mRNA and stable RNA. Nucleic Acids Res. 34, 659-666 (2006). https://doi.org/10.1093/nar/gkj472 PMid:16452296 PMCid:PMC1360286
|
|
|
101. Py, B., Higgins, C. F., Krisch, H. M. & Carpousis, A. J. A DEAD-box RNA helicase in the Escherichia coli RNA degradosome. Nature 381, 169-172 (1996).
https://doi.org/10.1038/381169a0 PMid:8610017
|
|
| |
|
102. Miczak, A., Kaberdin, V. R., Wei, C. L. & Lin-Chao, S. Proteins associated with RNase E in a multicomponent ribonucleolytic complex. Proc. Natl. Acad. Sci. U. S. A. 93, 3865-3869 (1996). https://doi.org/10.1073/pnas.93.9.3865 PMid:8632981 PMCid:PMC39450
|
|
|
103. Vanzo, N. F. et al. Ribonuclease E organizes the protein interactions in the Escherichia coli RNA degradosome. Genes Dev. 12, 2770-2781 (1998).
https://doi.org/10.1101/gad.12.17.2770 PMid:9732274 PMCid:PMC317140
|
|
|
104. Mohanty, B. K., Maples, V. F. & Kushner, S. R. The Sm-like protein Hfq regulates polyadenylation dependent mRNA decay in Escherichia coli. Mol. Microbiol. 54, 905-920 (2004). https://doi.org/10.1111/j.1365-2958.2004.04337.x PMid:15522076
|
|
|
105. Lin, C. L., Wang, Y.-T., Yang, W.-Z., Hsiao, Y.-Y. & Yuan, H. S. Crystal structure of human polynucleotide phosphorylase: insights into its domain function in RNA binding and degradation. Nucleic Acids Res. 40, 4146-4157 (2012). https://doi.org/10.1093/nar/gkr1281 PMid:22210891 PMCid:PMC3351181
|
|
|
106. Hayakawa, H. & Sekiguchi, M. Human polynucleotide phosphorylase protein in response to oxidative stress. Biochemistry 45, 6749-6755 (2006).
https://doi.org/10.1021/bi052585l PMid:16716086
|
|
|
107. Chen, X. et al. An ortholog of the Ro autoantigen functions in 23S rRNA maturation in D. radiodurans. Genes Dev. 21, 1328-1339 (2007). https://doi.org/10.1101/gad.1548207 PMid:17510283 PMCid:PMC1877746
|
|
|
108. Henry, A., Shanks, J., van Hoof, A. & Rosenzweig, J. A. The Yersinia pseudotuberculosis degradosome is required for oxidative stress, while its PNPase subunit plays a degradosome-independent role in cold growth. FEMS Microbiol. Lett. 336, 139-147 (2012). https://doi.org/10.1111/j.1574-6968.12000.x
PMid:23082859 PMCid:PMC5832447
|
|
|
109. Chen, H.-W., Koehler, C. M. & Teitell, M. A. Human polynucleotide phosphorylase: location matters. Trends Cell Biol. 17, 600-608 (2007).
https://doi.org/10.1016/j.tcb.2007.09.006 PMid:17983748
|
|
|
110. Malla, S. Unraveling the molecular mechanism of human polynucleotide phosphorylase (hPNPase) in controlling oxidized RNA. (Florida Atlantic University, 2019).
|
|
|
111. Szczesny, R. J. et al. Human mitochondrial RNA turnover caught in flagranti: involvement of hSuv3p helicase in RNA surveillance. Nucleic Acids Res. 38, 279-298 (2010). https://doi.org/10.1093/nar/gkp903 PMid:19864255 PMCid:PMC2800237
|
|
|
112. Wang, D. D.-H., Shu, Z., Lieser, S. A., Chen, P.-L. & Lee, W.-H. Human mitochondrial SUV3 and polynucleotide phosphorylase form a 330-kDa heteropentamer to cooperatively degrade double-stranded RNA with a 3'-to-5' directionality. J. Biol. Chem. 284, 20812-20821 (2009). https://doi.org/10.1074/jbc.M109.009605 PMid:19509288 PMCid:PMC2742846
|
|
|
113. Malla, S. & Li, Z. Functions of conserved domains of human polynucleotide phosphorylase on RNA oxidation. Insights Biomed Res 3, 62-67 (2019).
https://doi.org/10.36959/584/448 PMid:32123871 PMCid:PMC7051052
|
|
|
114. Orr, A. A. et al. A high-throughput and rapid computational method for screening of RNA post-transcriptional modifications that can be recognized by target proteins. Methods 143, 34-47 (2018). https://doi.org/10.1016/j.ymeth.2018.01.015 PMid:29408626
|
|
|
115. Gonzalez-Rivera, J. C. et al. Computational evolution of an RNA-binding protein towards enhanced oxidized-RNA binding. Comput. Struct. Biotechnol. J. 18, 137-152 (2020). https://doi.org/10.1016/j.csbj.2019.12.003 PMid:31988703 PMCid:PMC6965710
|
|
|
116. Cameron, T. A., Matz, L. M. & De Lay, N. R. Polynucleotide phosphorylase: Not merely an RNase but a pivotal post-transcriptional regulator. PLoS Genet. 14, e1007654 (2018). https://doi.org/10.1371/journal.pgen.1007654 PMid:30307990 PMCid:PMC6181284
|
|
|
117. Hardwick, S. W., Gubbey, T., Hug, I., Jenal, U. & Luisi, B. F. Crystal structure of Caulobacter crescentus polynucleotide phosphorylase reveals a mechanism of RNA substrate channelling and RNA degradosome assembly. Open Biol. 2, 120028 (2012). https://doi.org/10.1098/rsob.120028 PMid:22724061 PMCid:PMC3376730
|
|
|
118. Ohno, M. et al. A genome-wide distribution of 8-oxoguanine correlates with the preferred regions for recombination and single nucleotide polymorphism in the human genome. Genome Res. 16, 567-575 (2006). https://doi.org/10.1101/gr.4769606 PMid:16651663 PMCid:PMC1457041
|
|
|
119. Yoshihara, M., Jiang, L., Akatsuka, S., Suyama, M. & Toyokuni, S. Genome-wide profiling of 8-oxoguanine reveals its association with spatial positioning in nucleus. DNA Res. 21, 603-612 (2014). https://doi.org/10.1093/dnares/dsu023 PMid:25008760 PMCid:PMC4263294
|
|
|
120. Amente, S. et al. Genome-wide mapping of 8-oxo-7,8-dihydro-2'-deoxyguanosine reveals accumulation of oxidatively-generated damage at DNA replication origins within transcribed long genes of mammalian cells. Nucleic Acids Res. 47, 221-236 (2019). https://doi.org/10.1093/nar/gky1152 PMid:30462294 PMCid:PMC6326803
|
|
|
121. Ding, Y., Fleming, A. M. & Burrows, C. J. Sequencing the mouse genome for the oxidatively modified base 8-oxo-7,8-dihydroguanine by OG-Seq. J. Am. Chem. Soc. 139, 2569-2572 (2017). https://doi.org/10.1021/jacs.6b12604 PMid:28150947 PMCid:PMC5440228
|
|
|
122. Poetsch, A. R., Boulton, S. J. & Luscombe, N. M. Genomic landscape of oxidative DNA damage and repair reveals regioselective protection from mutagenesis. Genome Biol. 19, 215 (2018). https://doi.org/10.1186/s13059-018-1582-2 PMid:30526646 PMCid:PMC6284305
|
|
|
123. Tang, F. et al. Location analysis of 8-oxo-7,8-dihydroguanine in DNA by polymerase-mediated differential coding. Chem. Sci. 10, 4272-4281 (2019).
https://doi.org/10.1039/C8SC04946G PMid:31015952 PMCid:PMC6460952
|
|
|
124. McKinlay, A., Gerard, W. & Fields, S. Global analysis of RNA oxidation in Saccharomyces cerevisiae. Biotechniques 52, 109-111 (2012).
https://doi.org/10.2144/000113801 PMid:22313409 PMCid:PMC3339438
125. Gonzalez-Rivera, J. C. et al. Post-transcriptional air pollution oxidation to the cholesterol biosynthesis pathway promotes pulmonary stress phenotypes. Commun. Biol. 3, 392 (2020). https://doi.org/10.1038/s42003-020-01118-6 PMid:32699268 PMCid:PMC7376215
|
|
|