Abstract Lysosomes, membrane-bound organelles, play important roles in cellular processes including endocytosis, phagocytosis, and autophagy. Lysosomes maintain cellular homeostasis by generating a highly acidic environment of pH 4.5 – 5.0 and by housing hydrolytic enzymes that degrade engulfed biomolecules. Impairment of lysosomal function, especially in its acidification, is a driving force in the pathogenesis of diseases including neurodegeneration, cancer, metabolic disorders, and infectious diseases. Therefore, lysosomal pH is an attractive and targetable site for therapeutic intervention. Currently, there is a dearth of strategies or materials available to specifically modulate lysosomal acidification. This review focuses on the key aspects of how lysosomal pH is implicated in various diseases and discusses design strategies and molecular or nanoscale agents for lysosomal pH modulation, with the ultimate goal of developing novel therapeutic solutions.
Introduction
Lysosomes are membrane-bound vesicles that play key roles in the degradation and recycling of extracellular and intracellular materials, hence maintaining crucial cellular homeostasis and immune functions. Lysosomes contain numerous hydrolytic enzymes that degrade biological polymers such as proteins, lipids, nucleic acids and polysaccharides.(1,2) Lysosome function is tightly regulated by pH. Lysosomes are gaining increased attention for their role in cellular processes such as nutrient sensing, energy metabolism, as well as autophagy. A detailed discussion of these processes have been reviewed elsewhere3). Basal lysosomal pH is pH 4.5–5.0 with a buffering capacity of 19±6mM/pH unit(14), and multiple channels including chloride channels (e.g. CLC-7, CLC-6, CLC-3), calcium channels (e.g. TRPML1) and V-ATPase5), regulate this maintenance.
Dysregulation of lysosomal acidification, and defects in lysosome-organelle fusion lead to impairment of endocytic function, autophagic degradation, macromolecules biogenesis and transport3, and are observed in proteinopathic neurodegenerative diseases, metabolic disorders, and immunological diseases. Lysosomal acidification dysfunction also affects the activity of other organelles, such as mitochondria, leading to increased production of reactive oxygen species and inflammatory cytokines(6,7), thereby contributing to the pathogenesis of inflammatory diseases, cancer, and infectious diseases. Therefore, modulation of lysosomal acidification or pH is an increasingly important therapeutic target for disease management. In this review, we discuss the significance behind lysosomal pH modulation in diseases, with a focus on the major material design considerations for developing novel agents that specifically modulate lysosomal pH.
Lysosomal pH role and implication in disease pathogenesis
Lysosomes are the terminal degradative site/organelle for various cellular processes, including autophagy, which is an essential quality control machinery in the cells that degrade intracellular materials. Enhanced activation of the lysosome-autophagy process is being recognized as a driving force in the progression of numerous cancers, as it enables efficient nutrient scavenging and growth in nutrient-poor microenvironments8). Furthermore, the inhibition of lysosome-autophagy pathway leads to an increase in cellular apoptosis in tumors9). In pancreatic and lung adenocarcinomas, a lysosome mediated autophagic process systematically recycles cellular components, which provides nutrients for tumor growth in nutrient-deprived microenvironments. For instance, pancreatic ductal adenocarcinoma cells obtain essential amino acids and nucleotides from the lysosome-autophagy process when there is limited extracellular supply of these nutrients8). Small molecules such as chloroquine (CQ), hydroxychloroquine (HCQ) and its derivatives inhibit lysosomal acidification through elevating lysosomal pH, thereby reducing autophagic degradation, and are effective in reducing cancer progression in refractory multiple myeloma, glioblastoma, melanoma, and breast cancer models(10-14). Lysosome-autophagy processes are also implicated in the pathogenesis of viral diseases such as the human immunodeficiency virus (HIV) and the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). In dendritic cells, HIV inhibits lysosomal acidification and decreases lysosomal enzyme (e.g., cathepsins) activity. This effect impairs the digestion of viral particles, and reduces the ability of dendritic cells to inhibit antigen processing presentation to T cells(15). Additionally, the inhibition of lysosomal acidification with lysosomal V-ATPase inhibitor, Bafilomycin A1, increases the HIV-1 SF2 viral strain infectivity by close to 50 times(16). SARS-CoV-2 viral particles bind to ACE2 receptor and localize to the endolysosomal system, where the low pH environment cleaves the viral proteins and releases the viral RNA(17). Treatment with E64d cysteine protease inhibitor, which blocks lysosomal enzymatic function, and Bafilomycin A1, which elevates lysosomal pH, blocks SARS-CoV-2 S-protein-mediated endolysosomal entry(17,18). In clinical trials, where SARS-CoV-2 patients have been treated with HCQ, moderate responses in reducing SARS-CoV-2 viral load are seen(19). In autoimmune diseases such as lupus, significant lysosomal pH elevation is present both in MRL/lpr mice (i.e., lupus disease mouse model) and splenic B cells compared to normal controls. This lysosomal pH elevation results in the reduction of lysosomal cathepsins and membrane proteins activity, inhibits elimination of immune complexes that accumulate as a result of deficits in complement, as well as reduces expression of scavenger receptors and increases expression of Fcγ receptors(20), which initiate tissue inflammation through the release of harmful cytokines and chemokines(21). In Parkinson’s disease (PD) patients carrying mutations in the SNCA gene, which encodes for the presynaptic protein α-synuclein, formation of insoluble α-synuclein-containing aggregates disrupts the autophagy process, and inhibits lysosomal acidification(22,23). Similarly, in 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP) or 6-hydroxydopamine (6-OHDA)) induced experimental PD model, the exposure of neuroendocrine cells (e.g., PC-12 cells) to these neurotoxins results in mitochondria dysfunction, along with lysosomal pH elevation(24,25). Alzheimer’s disease is characterized by intracellular aggregates of the tau protein and extracellular β-amyloid (Aβ) plaques. Lysosomal acidification dysfunction affords mutations in the gene encoding the presenilin-2 component of the γ-secretase enzyme, leading to accumulation of cleaved Aβ plaques(26,27). In Down syndrome neurodevelopmental disorder which can lead to early-onset Alzheimer's disease, elevation of β cleaved carboxy terminal fragment of APP (APP-βCTF) results in lysosomal pH elevation and cellular dysfunction(28). Retinal pigmented epithelial (RPE) cells degrade both intra- and extracellular debris generated by autophagic degradation as well as phagocytosed photoreceptor outer segments. Chronic exposure of RPE cells to N-retinylidene-N-retinylethinolamide (A2E), a byproduct of the visual phototransduction cycle, affords de-regulation of lysosomal acidification, autophagy inhibition and accumulation of cellular waste, contributing to the pathogenesis of age-related macular degeneration(29-32). In metabolic disorders such as type II diabetes, pancreatic β-cells that have been chronically exposed to fatty acids (e.g., palmitic and oleic acids) show lysosomal acidification dysfunction, along with decreased mitochondrial activity and insulin sensitivity(33-35). Likewise, hepatocytes and cardiomyocytes exposed to high levels of fatty acids exhibit disrupted lysosomal acidification and cellular dysfunction(36,37). In sum, lysosomal pH plays a key role in disease pathogenesis, and it is imperative to design agents that specifically modulate lysosomal pH and rescue the associated cellular function impairments.
Biomaterial design strategies for modulating lysosomal pH
Currently, there are limited agents that specifically target lysosomal pH. The following design components or modules should be considered when synthesizing a novel material or agent to modulate lysosomal pH: 1) a lysosome localizing signal/motif or a delivery mechanism that enables efficient lysosome targeting; 2) a stimuli responsive biodegradable linker that affords degradation of the material within the lysosomal environment; and, 3) a liberated functional group from the degradation process, which is basic and increases lysosomal pH, or acidic and decreases the lysosomal pH. These design strategies are summarized in Figure 1.
Mechanism to ensure efficient targeting to the lysosomes
Key to this strategy is the targeting of materials – molecular or nanosized to the lysosome. Specific lysosome targeting is important because unspecific organelle targeting can potentially affect other organelles, such as the mitochondria, which in turn crosstalk and modulate lysosomal function as well6,38,39). One approach for lysosomal specific targeting is through the conjugation of lysosomal membrane targeting motifs, which enable specific uptake via receptor mediated endocytosis. In general, there are two classes of motifs for lysosomal targeting: A) tyrosine-based (Y) motif with YXXÏ• consensus sequence, where X can be any amino acid and Ï• is amino acid with bulky hydrophobic side chain that mainly targets the transferrin receptor, LAMP-1 or CD1(40), and B) dileucine-based motif with either a [DE]XXXL[LI] (i.e., the square brackets indicate alternatives) or a DXXLL pattern(41-43). Additionally, the (aminoethyl)morpholine chemical group is a lysosome targeting motif(44,45). Alternatively, conjugation of a small molecule to the mannose-6-phosphate group or derivative affords lysosome accumulation as the sugar is recognized by the Golgi and subsequently transported to the endosomal/lysosomal system(46,47). For instance, the lysosome targeting chimeras (LYTACS), which composes of a target binding moiety (e.g., small molecule) linked to mannose-6-phosphonate (M6Pn), a CI-M6PR binding ligand, allow for specific lysosome localization(48). Ahn et al. further modified the LYTACs design to be liver cell specific, through conjugation of an asialoglycoprotein receptor, which recognizes glycoproteins bearing N-acetylgalactosamine or galactose ligands present on liver cells(49). To target specifically to brain cells, blood barrier targeting peptides are also used(50).
A second general strategy is to use nanosized particles, of diameters ranging from 25 – 200 nm, which rapidly uptake into cells and localize to subcellular organelles (e.g., lysosomes) via endocytosis(51,52). The sizes and shapes of these nanoparticles are finely tuned by varying the material, synthesis methods, and types of surface ligand, which also influence their targeting and localization efficiency(53). To improve the lysosomal targeting and localization efficiency of hydrophilic CQ and HCQ molecules (Figure 2), encapsulation of these small molecules in poly (lactic acid) nanoparticles affords higher efficacy in reducing Herpes simplex virus type 1 infections in VeroE6 cells compared to either using CQ or HCQ alone(54,55). In another instance, CQ encapsulated poly (lactic-co-glycolic) acid polyester (PLGA) nanoparticles increase autophagy inhibition in A549 multidrug resistance cells(56). HCQ loaded hollow mesoporous silica nanoparticles localize to lysosomes and improve autophagic inhibition and therapeutic efficacy in colon tumor tissues(57). HCQ loaded liposomes decorated with a pH-sensitive TH-RGD targeting peptide increase accumulation of HCQ in B16F10 tumor cells and lysosomes, leading to blockage of autophagic flux in tumor cells, along with reduced tumor growth(58). Finally, liposomal nanocarriers modified with octadecyl-rhodamine B, localize to lysosomes, suggesting that this is another approach to targeting lysosome accumulation(59,60).
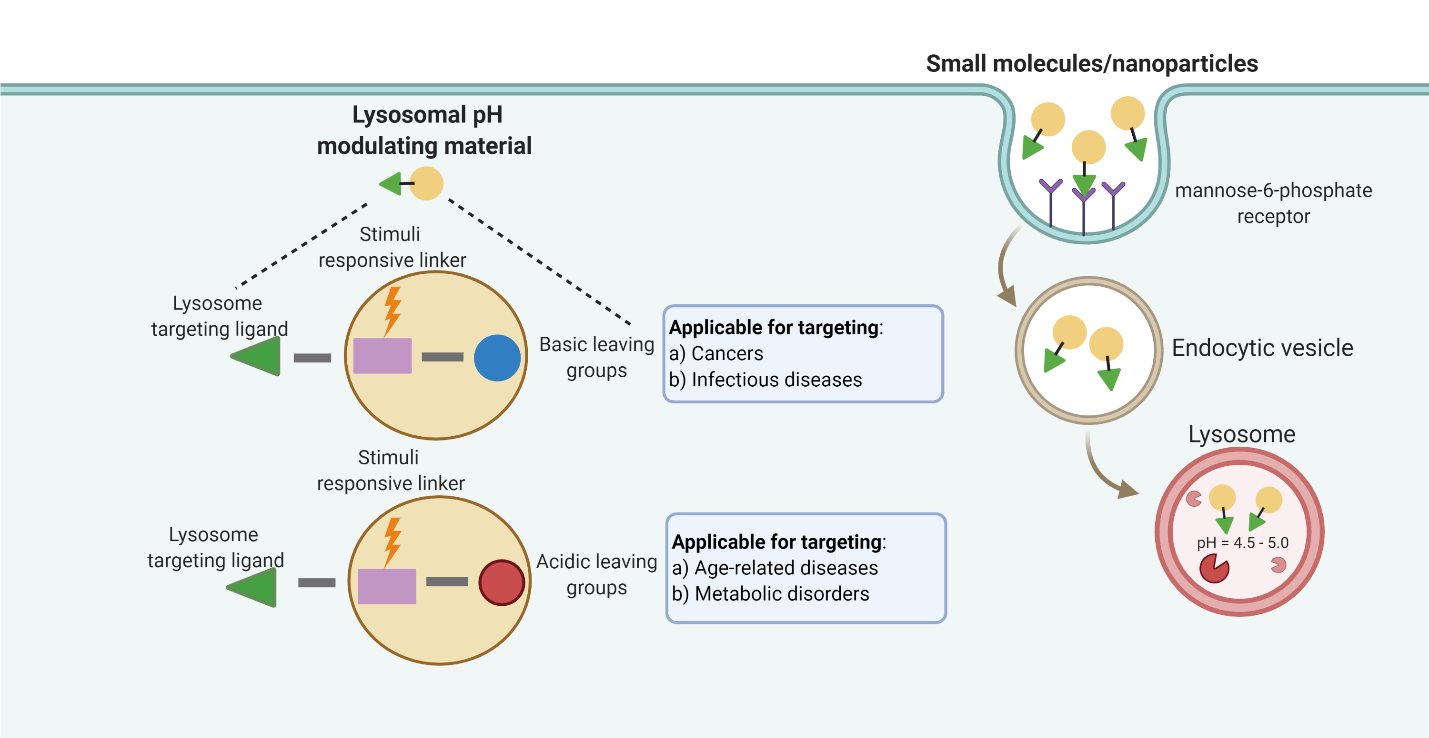
Figure 1. Schematic showing the material design strategies, including a cell membrane targetable ligand, which allows for lysosomal localization via receptor mediated endocytosis, a stimuli responsive linker that is degradable, and a liberated group that can modulate lysosomal pH. This schematic is created with BioRender.com.
Stimuli responsive biodegradable linkages
Polymers with weakly acidic or basic residues are often utilized as pH-responsive polymers. This group of polymers includes polyesters, polyanhydrides, polycarbonates and others, and their properties and synthesis procedures have been reviewed elsewhere(61). Polyesters can be readily hydrolyzed in mild aqueous acid to release carboxylic acids groups. For instance, poly (lactic-co-glycolic) acid polyester (PLGA) based nanoparticle, with an ester linkage, release carboxylic acid upon contact with the low lysosomal pH environment (i.e., pH 6.0), and further lower lysosomal pH in MPTP-induced PD(24,25), type II diabetes(62) and age-related macular degeneration(31)disease cell models. Polyanhydrides also degrade into component carboxylic acids; however, polyanhydrides generally have a lower degree of dissociation and do not change pH value as much as polyesters within similar time frame of degradation(63,64). The lysosomal environment contains enzymes, such as cathepsins, which hydrolyze and degrade chemical linkages, hence several research groups are investigating cathepsin B enzyme specific degradable linkages that enable specific compound release upon enzymatic cleavage(65). Apart from leveraging the lysosomal environment as a stimuli/trigger, light-activated cleavable linkages are also employed to release acids to modulate lysosomal pH. Using a UV-light (e.g., 365 nm) labile biodegradable linkage group, 1-(2-nitrophenyl)ethan-1-ol, Trudeau et al. reduce lysosomal pH in pancreatic β-cells under chronic exposure to palmitic acid, through the release of carboxylic acid groups from a nanoparticle(34,35). Other externally applied stimuli include near-infrared (750 nm) responsive cleavable linkages, which enable higher tissue penetration depth compared to UV light, and are of utility for applications in in vivo models(66). Such near-infrared systems are worthy to explore for this application.
Liberated basic/acidic functional groups increase/decrease lysosomal pH
To either increase or decrease lysosomal pH, the component group released upon cleavage by the acidic lysosomal environment or enzyme should be either a base or an acid. The pKa of the released component should be higher than the basal lysosomal pH (4.5 – 5.0) for pH elevation, or lower than the basal level for pH reduction. Small molecules such as hydroxychloroquine (HCQ) (pKas of 4.0, 8.3 and 9.7), and chloroquine (CQ) (pKas of 4.0, 8.4 and 10.2), elevate lysosomal pH, due to the presence of basic side chains, which allows it to act as weak bases(67). In African green monkey kidney VeroE6 cells infected with the SARS-CoV-2 virus, the addition of either CQ or HCQ inhibits lysosomal acidification, along with a reduction in virus multiplicities of infection(68). Analogues of CQ/HCQ, such as Nitazoxanide (pKa 8.3) and ROC-325 (pKa 8.3)(69,70), also inhibit lysosomal acidification, and reduce glioma and acute myeloid leukemia cell viability and proliferation, respectively (Figure 2)(69-71). Small molecules IITZ-01 (pKas 4.7, 5.4, 11.54, 12.54 13.7, 54.88) and IITZ-02 (pKa 4.75, 5.42, 11.56, 12.65, 14.49), which are benzimidazole containing two s-triazine analogs, are basic and de-acidify lysosomes and inhibit tumor growth in triple negative breast cancer cellular and mouse models(14). Comprising of two basic pyrrole groups, obatoclax mesylate (pKas 4.68, 13.97) also elevate lysosomal pH in ovarian cancer models, thereby contributing to cell death(72,73). Contrary to releasing basic groups to elevate lysosomal pH, Hong et al. synthesized compounds based on lysosome-targeting fluorescent anion transporters derived from coumarins, trifluoromethylated arylsquaramides and morpholines, which elevate lysosomal pH through increasing chloride ions efflux out of the lysosomes(74).
In contrast, the number of compounds that lower lysosomal pH is substantially smaller. Materials that release acidic components to lower the lysosomal pH include the polyester, poly (lactic-co-glycolic acid) (PLGA). PLGA degrades in an acidic environment to release lactic and glycolic component carboxylic acids with pKas of 3.86 and 3.83 respectively6(62), resulting in lysosomal pH reduction in various disease models with elevated lysosomal pH(24,25,31,75,76). The chemical structures for these agents or materials mentioned above are shown in Figure 2.
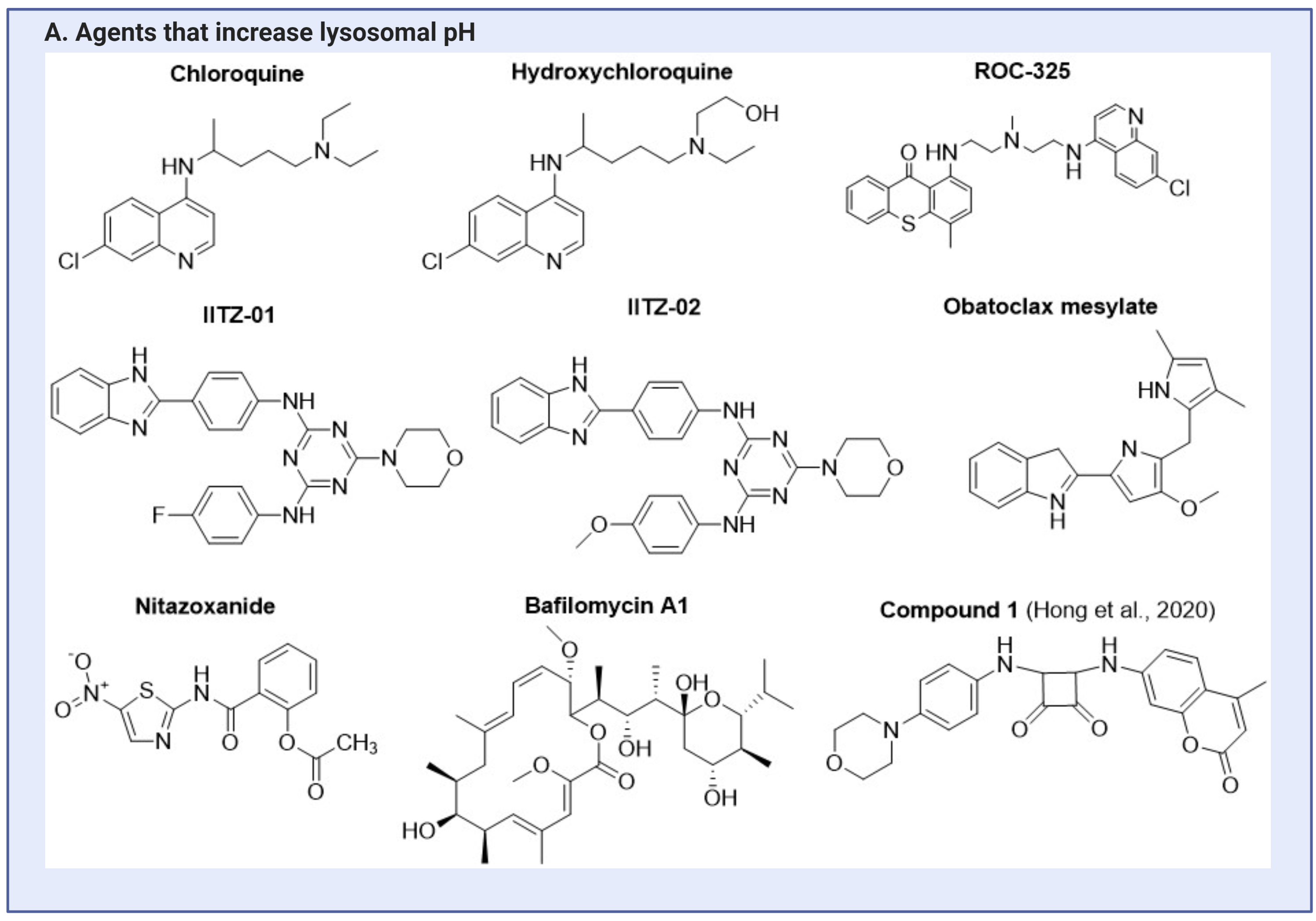
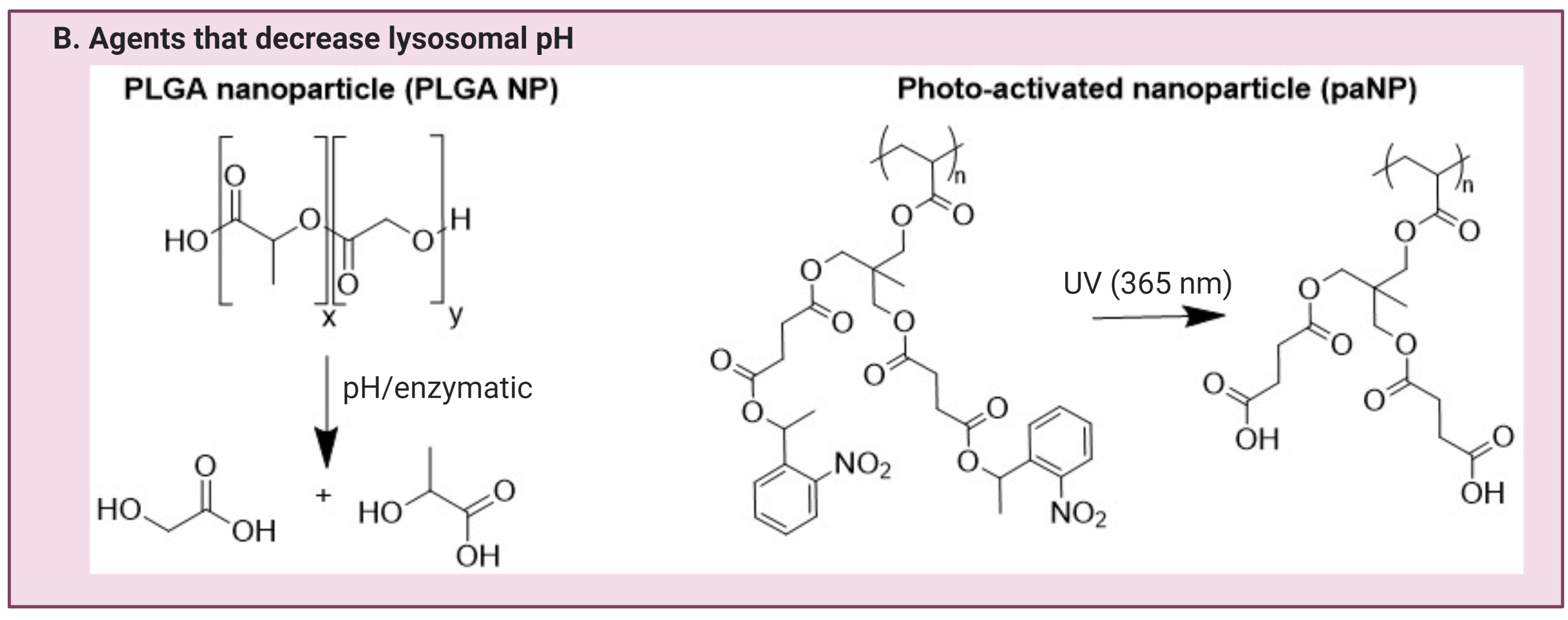
Figure 2. Agents that are currently being investigated for lysosomal pH modulating applications. This can be categorized into agents that can either A) elevate lysosomal pH, or B) lower lysosomal pH. This figure is created with BioRender.com.
Conclusion
Lysosomal pH is an important modulator of various cellular processes. Herein, we review the significance of lysosomal acidification in disease pathogenesis, as well as material designs and strategies to ensure efficient lysosomal targeting and modulation of lysosomal pH. These strategies include the presence of a lysosome targeting motif, a stimuli-responsive degradable linker, either acidic or basic functional groups, and the potential to encapsulate or deliver active agents in the form of nanoparticles. Depending on the specific applications, some or all the design strategies are applicable to the materials in development. To ensure that the change in cellular function is solely due to a change in lysosomal pH, it is also important to design experiments with proper controls, such as through the addition of lysosomal V-ATPase inhibitor Bafilomycin A1, and lysosomal protease inhibitor, E64d. Since lysosome function can also affect other organelle functions (e.g., mitochondria function), it is prudent to assess the function of these organelles, along with lysosomal functional changes. Finally, evaluation of these materials or agents in in vivo models to determine efficacy and pharmacokinetic/pharmacodynamic parameters is critical to pre-clinical development. With proper experimental controls and designs, we will gain a better mechanistic understanding of the effect of lysosomal pH on downstream cellular processes and insights on optimizing agents that control lysosomal acidification to rescue cellular dysfunctions. Our vision is to translate these materials or concepts into therapeutic agents.
Author contributions
JLZ conceptualized the idea and wrote the manuscript. OSS and MWG have edited and consented to the final version of the manuscript.
Acknowledgements
This work was funded by National Institutes of Health R21 grant NIH AG063373 and AG060456, BU Nanotechnology center at Boston University.
The authors declare no competing financial interest.
References
|
1. Wang, F., Gómez-Sintes, R. & Boya, P. Lysosomal membrane permeabilization and cell death. Traffic 19, 918-931 (2018).
https://doi.org/10.1111/tra.12613 PMid:30125440
|
| |
|
2. De Duve, C., Pressman, B. C., Gianetto, R., Wattiaux, R. & Appelmans, F. Tissue fractionation studies. 6. Intracellular distribution patterns of enzymes in rat-liver tissue. Biochem. J. 60, 604-617 (1955). https://doi.org/10.1042/bj0600604 PMid:13249955 PMCid:PMC1216159
|
|
| |
|
3. Lawrence, R. E. & Zoncu, R. The lysosome as a cellular centre for signalling, metabolism and quality control. Nat. Cell Biol. 21, 133-142 (2019).
https://doi.org/10.1038/s41556-018-0244-7 PMid:30602725
|
|
| |
|
4. Gekle, M. & Silbernagl, S. Comparison of the buffer capacity of endocytotic vesicles, lysosomes and cytoplasm in cells derived from the proximal tubule of the kidney (opossum kidney cells). Pflugers Arch. 429, 452-454 (1995). https://doi.org/10.1007/BF00374165 PMid:7761271
|
|
|
|
|
5. Mindell, J. A. Lysosomal acidification mechanisms. Annu. Rev. Physiol. 74, 69-86 (2012).https://doi.org/10.1146/annurev-physiol-012110-142317 PMid:22335796
|
|
| |
|
6. Deus, C. M., Yambire, K. F., Oliveira, P. J. & Raimundo, N. Mitochondria-Lysosome Crosstalk: From Physiology to Neurodegeneration. Trends Mol. Med. 26, 71-88 (2020). https://doi.org/10.1016/j.molmed.2019.10.009 PMid:31791731
|
|
| |
|
7. Yambire, K. F. et al. Impaired lysosomal acidification triggers iron deficiency and inflammation in vivo. Elife 8, e51031 (2019). https://doi.org/10.7554/eLife.51031 PMid:31793879 PMCid:PMC6917501
|
|
| |
|
8. White, E. The role for autophagy in cancer. J. Clin. Invest. 125, 42-46 (2015). https://doi.org/10.1172/JCI73941 PMid:25654549 PMCid:PMC4382247
|
|
| |
|
9. Desantis, V. et al. Autophagy: A New Mechanism of Prosurvival and Drug Resistance in Multiple Myeloma. Transl. Oncol. 11, 1350-1357 (2018). https://doi.org/10.1016/j.tranon.2018.08.014 PMid:30196237 PMCid:PMC6132177
|
|
| |
|
10. Mahalingam, D. et al. Combined autophagy and HDAC inhibition: a phase I safety, tolerability, pharmacokinetic, and pharmacodynamic analysis of hydroxychloroquine in combination with the HDAC inhibitor vorinostat in patients with advanced solid tumors. Autophagy 10, 1403-1414 (2014). https://doi.org/10.4161/auto.29231 PMid:24991835 PMCid:PMC4203517
|
|
| |
|
11. Rangwala, R. et al. Combined MTOR and autophagy inhibition: phase I trial of hydroxychloroquine and temsirolimus in patients with advanced solid tumors and melanoma. Autophagy 10, 1391-1402 (2014). https://doi.org/10.4161/auto.29119 https://doi.org/10.4161/auto.29118
|
|
| |
|
12. Rangwala, R. et al. Phase I trial of hydroxychloroquine with dose-intense temozolomide in patients with advanced solid tumors and melanoma. Autophagy 10, 1369-1379 (2014). https://doi.org/10.4161/auto.29119 https://doi.org/10.4161/auto.29118 PMid:24991839 PMCid:PMC4203514
|
|
| |
|
13. Rosenfeld, M. R. et al. A phase I/II trial of hydroxychloroquine in conjunction with radiation therapy and concurrent and adjuvant temozolomide in patients with newly diagnosed glioblastoma multiforme. Autophagy 10, 1359-1368 (2014). https://doi.org/10.4161/auto.28984 PMid:24991840 PMCid:PMC4203513
|
|
| |
|
14. Guntuku, L. et al. IITZ-01, a novel potent lysosomotropic autophagy inhibitor, has single-agent antitumor efficacy in triple-negative breast cancer in vitro and in vivo. Oncogene 38, 581-595 (2019). https://doi.org/10.1038/s41388-018-0446-2 PMid:30166591
|
|
| |
|
15. Zhou, D., Kang, K. H. & Spector, S. A. Production of interferon α by human immunodeficiency virus type 1 in human plasmacytoid dendritic cells is dependent on induction of autophagy. J. Infect. Dis. 205, 1258-1267 (2012). https://doi.org/10.1093/infdis/jis187 PMid:22396599 PMCid:PMC3308911
|
|
| |
|
16. Fredericksen, B. L., Wei, B. L., Yao, J., Luo, T. & Garcia, J. V. Inhibition of endosomal/lysosomal degradation increases the infectivity of human immunodeficiency virus. J. Virol. 76, 11440-11446 (2002). https://doi.org/10.1128/JVI.76.22.11440-11446.2002 PMid:12388705 PMCid:PMC136743
|
|
| |
|
17. Hoffmann, M. et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 181, 271-280.e8 (2020). https://doi.org/10.1016/j.cell.2020.02.052 PMid:32142651 PMCid:PMC7102627
|
|
| |
|
18. Blaess, M., Kaiser, L., Sauer, M., Csuk, R. & Deigner, H.-P. COVID-19/SARS-CoV-2 Infection: Lysosomes and Lysosomotropism Implicate New Treatment Strategies and Personal Risks. Int. J. Mol. Sci. 21, 4953 (2020). https://doi.org/10.3390/ijms21144953 PMid:32668803 PMCid:PMC7404102
|
|
| |
|
19. Gautret, P. et al. Hydroxychloroquine and azithromycin as a treatment of COVID-19: results of an open-label non-randomized clinical trial. Int. J. Antimicrob. Agents 105949 (2020) doi:10.1016/j.ijantimicag.2020.105949. https://doi.org/10.1016/j.ijantimicag.2020.105949 PMid:32205204 PMCid:PMC7102549
|
|
| |
|
20. Pickering, M. C., Botto, M., Taylor, P. R., Lachmann, P. J. & Walport, M. J. Systemic lupus erythematosus, complement deficiency, and apoptosis. Adv. Immunol. 76, 227-324 (2000). https://doi.org/10.1016/S0065-2776(01)76021-X
|
|
| |
|
21. Monteith, A. J. et al. Defects in lysosomal maturation facilitate the activation of innate sensors in systemic lupus erythematosus. Proc. Natl. Acad. Sci. U. S. A. 113, E2142-51 (2016). https://doi.org/10.1073/pnas.1513943113 PMid:27035940 PMCid:PMC4839468
|
|
| |
|
22. Winslow, A. R. et al. α-Synuclein impairs macroautophagy: implications for Parkinson's disease. J. Cell Biol. 190, 1023-1037 (2010). https://doi.org/10.1083/jcb.201003122 PMid:20855506 PMCid:PMC3101586
|
|
| |
|
23. Pan, T., Kondo, S., Le, W. & Jankovic, J. The role of autophagy-lysosome pathway in neurodegeneration associated with Parkinson's disease. Brain 131, 1969-1978 (2008). https://doi.org/10.1093/brain/awm318 PMid:18187492
|
|
| |
|
24. Zeng, J., Martin, A., Han, X., Shirihai, O. S. O. S. & Grinstaff, M. W. M. W. Biodegradable PLGA Nanoparticles Restore Lysosomal Acidity and Protect Neural PC-12 Cells against Mitochondrial Toxicity. Ind. Eng. Chem. Res. 58, 13910-13917 (2019). https://doi.org/10.1021/acs.iecr.9b02003
|
|
| |
|
25. Bourdenx, M. et al. Nanoparticles restore lysosomal acidification defects: Implications for Parkinson and other lysosomal-related diseases. Autophagy 12, 472-483 (2016). https://doi.org/10.1080/15548627.2015.1136769 PMid:26761717 PMCid:PMC4835967
|
|
| |
|
26. Cataldo, A. M. et al. App gene dosage modulates endosomal abnormalities of Alzheimer's disease in a segmental trisomy 16 mouse model of down syndrome. J. Neurosci. 23, 6788-6792 (2003). https://doi.org/10.1523/JNEUROSCI.23-17-06788.2003 PMid:12890772 PMCid:PMC6740714
|
|
| |
|
27. Li, X. et al. Discovery of nitazoxanide-based derivatives as autophagy activators for the treatment of Alzheimer's disease. Acta Pharm. Sin. B 10, 646-666 (2020). https://doi.org/10.1016/j.apsb.2019.07.006 PMid:32322468 PMCid:PMC7161708
|
|
| |
|
28. Jiang, Y. et al. Lysosomal Dysfunction in Down Syndrome Is APP-Dependent and Mediated by APP-βCTF (C99). J. Neurosci. 39, 5255 LP - 5268 (2019).
https://doi.org/10.1523/JNEUROSCI.0578-19.2019 PMid:31043483 PMCid:PMC6607756
|
|
| |
|
29. Liu, J. et al. Restoration of lysosomal pH in RPE cells from cultured human and ABCA4(-/-) mice: pharmacologic approaches and functional recovery. Invest. Ophthalmol. Vis. Sci. 49, 772-780 (2008). https://doi.org/10.1167/iovs.07-0675 PMid:18235027 PMCid:PMC2279299
|
|
| |
|
30. Lu, W. et al. The P2Y12 Receptor Antagonist Ticagrelor Reduces Lysosomal pH and Autofluorescence in Retinal Pigmented Epithelial Cells From the ABCA4-/- Mouse Model of Retinal Degeneration. Front. Pharmacol. 9, 242 (2018). https://doi.org/10.3389/fphar.2018.00242 PMid:29725296 PMCid:PMC5917064
|
|
| |
|
31. Baltazar, G. C. et al. Acidic nanoparticles are trafficked to lysosomes and restore an acidic lysosomal pH and degradative function to compromised ARPE-19 cells. PLoS One 7, e49635 (2012). https://doi.org/10.1371/journal.pone.0049635 PMid:23272048 PMCid:PMC3525582
|
|
| |
|
32. Guha, S., Liu, J., Baltazar, G., Laties, A. M. & Mitchell, C. H. Rescue of compromised lysosomes enhances degradation of photoreceptor outer segments and reduces lipofuscin-like autofluorescence in retinal pigmented epithelial cells. Adv. Exp. Med. Biol. 801, 105-111 (2014). https://doi.org/10.1007/978-1-4614-3209-8_14 PMid:24664687 PMCid:PMC4163923
|
|
| |
|
33. Las, G., Serada, S. B., Wikstrom, J. D., Twig, G. & Shirihai, O. S. Fatty acids suppress autophagic turnover in β-cells. J. Biol. Chem. 286, 42534-42544 (2011). https://doi.org/10.1074/jbc.M111.242412 PMid:21859708 PMCid:PMC3234912
|
|
| |
|
34. Assali, E. A. et al. Nanoparticle-mediated lysosomal reacidification restores mitochondrial turnover and function in β cells under lipotoxicity. FASEB J. (2019) doi:10.1096/fj.201801292R. https://doi.org/10.1096/fj.201801292R PMid:30550357
|
|
| |
|
35. Trudeau, K. M. et al. Lysosome acidification by photoactivated nanoparticles restores autophagy under lipotoxicity. J. Cell Biol. 214, 25-34 (2016).
https://doi.org/10.1083/jcb.201511042 PMid:27377248 PMCid:PMC4932370
|
|
| |
|
36. Inami, Y. et al. Hepatic steatosis inhibits autophagic proteolysis via impairment of autophagosomal acidification and cathepsin expression. Biochem. Biophys. Res. Commun. 412, 618-25 (2011). https://doi.org/10.1016/j.bbrc.2011.08.012 PMid:21856284
|
|
| |
|
37. Liu, Y. et al. Palmitate-Induced Vacuolar-Type H(+)-ATPase Inhibition Feeds Forward Into Insulin Resistance and Contractile Dysfunction. Diabetes 66, 1521-1534 (2017). https://doi.org/10.2337/db16-0727 PMid:28302654
|
|
| |
|
38. Plotegher, N. & Duchen, M. R. Crosstalk between Lysosomes and Mitochondria in Parkinson's Disease . Frontiers in Cell and Developmental Biology vol. 5 110 (2017). https://doi.org/10.3389/fcell.2017.00110 PMid:29312935 PMCid:PMC5732996
|
|
| |
|
39. Guerra, F. et al. Synergistic Effect of Mitochondrial and Lysosomal Dysfunction in Parkinson's Disease. Cells 8, (2019). https://doi.org/10.3390/cells8050452
PMid:31091796 PMCid:PMC6563092
|
|
| |
|
40. Pandey, K. N. Small peptide recognition sequence for intracellular sorting. Curr. Opin. Biotechnol. 21, 611-620 (2010). https://doi.org/10.1016/j.copbio.2010.08.007 PMid:20817434 PMCid:PMC2997389
|
|
| |
|
41. Behnke, J., Eskelinen, E.-L., Saftig, P. & Schröder, B. Two dileucine motifs mediate late endosomal/lysosomal targeting of transmembrane protein 192 (TMEM192) and a C-terminal cysteine residue is responsible for disulfide bond formation in TMEM192 homodimers. Biochem. J. 434, 219-231 (2011).
https://doi.org/10.1042/BJ20101396 PMid:21143193
|
|
| |
|
42. Sakhrani, N. M. & Padh, H. Organelle targeting: third level of drug targeting. Drug Des. Devel. Ther. 7, 585-599 (2013). https://doi.org/10.2147/DDDT.S45614
PMid:23898223 PMCid:PMC3718765
|
|
| |
|
43. Contu, V. R. et al. Lysosomal targeting of SIDT2 via multiple YxxΦ motifs is required for SIDT2 function in the process of RNautophagy. J. Cell Sci. 130, 2843 LP - 2853 (2017). https://doi.org/10.1242/jcs.202481 PMid:28724756
|
|
| |
|
44. Yang, S. et al. Design of a simultaneous target and location-activatable fluorescent probe for visualizing hydrogen sulfide in lysosomes. Anal. Chem. 86, 7508-7515 (2014). https://doi.org/10.1021/ac501263d PMid:24975419
|
|
| |
|
45. Wan, Q., Chen, S., Shi, W., Li, L. & Ma, H. Lysosomal pH rise during heat shock monitored by a lysosome-targeting near-infrared ratiometric fluorescent probe. Angew. Chem. Int. Ed. Engl. 53, 10916-10920 (2014). https://doi.org/10.1002/anie.201405742 PMid:25154475
|
|
| |
|
46. Coutinho, M. F., Prata, M. J. & Alves, S. Mannose-6-phosphate pathway: a review on its role in lysosomal function and dysfunction. Mol. Genet. Metab. 105, 542-550 (2012). https://doi.org/10.1016/j.ymgme.2011.12.012 PMid:22266136
|
|
| |
|
47. Louzoun-Zada, S., Jaber, Q. Z. & Fridman, M. Guiding Drugs to Target-Harboring Organelles: Stretching Drug-Delivery to a Higher Level of Resolution. Angew. Chemie Int. Ed. 58, 15584-15594 (2019). https://doi.org/10.1002/anie.201906284 PMid:31237741
|
|
| |
|
48. Banik, S. M. et al. Lysosome-targeting chimaeras for degradation of extracellular proteins. Nature 584, 291-297 (2020). https://doi.org/10.1038/s41586-020-2545-9 PMid:32728216
|
|
| |
|
49. Green Ahn, Steven Banik, Caitlyn L. Miller, Nicholas Riley, Jennifer R. Cochran, C. B. Lysosome Targeting Chimeras (LYTACs) That Engage a Liver-Specific Asialoglycoprotein Receptor for Targeted Protein Degradation. ChemRxiv (2020).
|
|
| |
|
50. McCully, M., Sanchez-Navarro, M., Teixido, M. & Giralt, E. Peptide Mediated Brain Delivery of Nano- and Submicroparticles: A Synergistic Approach. Curr. Pharm. Des. 24, 1366-1376 (2018). https://doi.org/10.2174/1381612824666171201115126 PMid:29205110 PMCid:PMC6110044
|
|
| |
|
51. Zhang, S., Li, J., Lykotrafitis, G., Bao, G. & Suresh, S. Size-Dependent Endocytosis of Nanoparticles. Adv. Mater. 21, 419-424 (2009). https://doi.org/10.1002/adma.200801393 PMid:19606281 PMCid:PMC2709876
|
|
| |
|
52. Rathore, B. et al. Nanomaterial designing strategies related to cell lysosome and their biomedical applications: A review. Biomaterials 211, 25-47 (2019). https://doi.org/10.1016/j.biomaterials.2019.05.002 PMid:31078050
|
|
| |
|
53. Caldorera-Moore, M., Guimard, N., Shi, L. & Roy, K. Designer nanoparticles: incorporating size, shape and triggered release into nanoscale drug carriers. Expert Opin. Drug Deliv. 7, 479-495 (2010). https://doi.org/10.1517/17425240903579971 PMid:20331355 PMCid:PMC2845970
|
|
| |
|
54. Lima, T. L. C. et al. Improving Encapsulation of Hydrophilic Chloroquine Diphosphate into Biodegradable Nanoparticles: A Promising Approach against Herpes Virus Simplex-1 Infection. Pharmaceutics 10, 255 (2018). https://doi.org/10.3390/pharmaceutics10040255 PMid:30513856 PMCid:PMC6320969
|
|
| |
|
55. Oller-Salvia, B., Sánchez-Navarro, M., Giralt, E. & Teixidó, M. Blood-brain barrier shuttle peptides: an emerging paradigm for brain delivery. Chem. Soc. Rev. 45, 4690-4707 (2016). https://doi.org/10.1039/C6CS00076B PMid:27188322
|
|
| |
|
56. Sun, J.-H. et al. Co-delivery nanoparticles of doxorubicin and chloroquine for improving the anti-cancer effect in vitro. Nanotechnology 30, 85101 (2018).
https://doi.org/10.1088/1361-6528/aaf51b PMid:30523865
|
|
| |
|
57. Li, Y. et al. Hydroxychloroquine-loaded hollow mesoporous silica nanoparticles for enhanced autophagy inhibition and radiation therapy. J. Control. Release 325, 100-110 (2020). https://doi.org/10.1016/j.jconrel.2020.06.025 PMid:32621826
|
|
| |
|
58. Wang, Y. et al. Significantly enhanced tumor cellular and lysosomal hydroxychloroquine delivery by smart liposomes for optimal autophagy inhibition and improved antitumor efficiency with liposomal doxorubicin. Autophagy 12, 949-962 (2016). https://doi.org/10.1080/15548627.2016.1162930 PMid:27123811 PMCid:PMC4922436
|
|
| |
|
59. Meerovich, I., Koshkaryev, A., Thekkedath, R. & Torchilin, V. P. Screening and Optimization of Ligand Conjugates for Lysosomal Targeting. Bioconjug. Chem. 22, 2271-2282 (2011). https://doi.org/10.1021/bc200336j PMid:21913714 PMCid:PMC3218248
|
|
| |
|
60. Torchilin, V. P. Next step in drug delivery: getting to individual organelles. Drug Deliv. Transl. Res. 2, 415-417 (2012). https://doi.org/10.1007/s13346-012-0102-2 PMid:23565351 PMCid:PMC3615566
|
|
| |
|
61. Kocak, G., Tuncer, C. & Bütün, V. pH-Responsive polymers. Polym. Chem. 8, 144-176 (2017). https://doi.org/10.1039/C6PY01872F
|
|
| |
|
62. Zeng, J., Shirihai, O. S. & Grinstaff, M. W. Degradable Nanoparticles Restore Lysosomal pH and Autophagic Flux in Lipotoxic Pancreatic Beta Cells. Adv. Healthc. Mater. 0, 1801511 (2019).https://doi.org/10.1002/adhm.201801511 PMid:30698920
|
|
| |
|
63. Chu, I.-M., Liu, T.-H. & Chen, Y.-R. Preparation and characterization of sustained release system based on polyanhydride microspheres with core/shell-like structures. J. Polym. Res. 26, 1 (2018). https://doi.org/10.1007/s10965-018-1657-5
|
|
| |
|
64. Carbone, A. L. & Uhrich, K. E. Design and Synthesis of Fast-Degrading Poly(anhydride-esters). Macromol. Rapid Commun. 30, 1021 (2009).https://doi.org/10.1002/marc.200900029 PMid:20161638 PMCid:PMC2818006
|
|
| |
|
65. Wang, Y. et al. Lysosome-Targeting Fluorogenic Probe for Cathepsin B Imaging in Living Cells. Anal. Chem. 88, 12403-12410 (2016).
https://doi.org/10.1021/acs.analchem.6b03717 PMid:28193055
|
|
| |
|
66. Luciano, M. P. et al. Chapter Eleven - A near-infrared light-mediated cleavable linker strategy using the heptamethine cyanine chromophore. in Chemical Tools for Imaging, Manipulating, and Tracking Biological Systems: Diverse Chemical, Optical and Bioorthogonal Methods (ed. Chenoweth, D. M. B. T.-M. in E.) vol. 641 245-275 (Academic Press, 2020). https://doi.org/10.1016/bs.mie.2020.04.043 PMid:32713525
|
|
| |
|
67. Schrezenmeier, E. & Dörner, T. Mechanisms of action of hydroxychloroquine and chloroquine: implications for rheumatology. Nat. Rev. Rheumatol. 16, 155-166 (2020). https://doi.org/10.1038/s41584-020-0372-x PMid:32034323
|
|
| |
|
68. Liu, J. et al. Hydroxychloroquine, a less toxic derivative of chloroquine, is effective in inhibiting SARS-CoV-2 infection in vitro. Cell Discov. 6, 16 (2020).
https://doi.org/10.1038/s41421-020-0156-0 PMid:32194981 PMCid:PMC7078228
|
|
| |
|
69. Wang, X. et al. Nitazoxanide, an antiprotozoal drug, inhibits late-stage autophagy and promotes ING1-induced cell cycle arrest in glioblastoma. Cell Death Dis. 9, 1032 (2018). https://doi.org/10.1038/s41419-018-1058-z PMid:30302016 PMCid:PMC6177448
|
|
| |
|
70. Jones, T. M., Espitia, C., Wang, W., Nawrocki, S. T. & Carew, J. S. Moving beyond hydroxychloroquine: the novel lysosomal autophagy inhibitor ROC-325 shows significant potential in preclinical studies. Cancer Commun. 39, 72 (2019). https://doi.org/10.1186/s40880-019-0418-0 PMid:31706349 PMCid:PMC6842502
|
|
| |
|
71. Nawrocki, S. T. et al. The novel autophagy inhibitor ROC-325 augments the antileukemic activity of azacitidine. Leukemia 33, 2971-2974 (2019).https://doi.org/10.1038/s41375-019-0529-2 PMid:31358855 PMCid:PMC7462348
|
|
| |
|
72. Stamelos, V. A. et al. The BH3 Mimetic Obatoclax Accumulates in Lysosomes and Causes Their Alkalinization. PLoS One 11, e0150696-e0150696 (2016). https://doi.org/10.1371/journal.pone.0150696 PMid:26950068 PMCid:PMC4780728
|
|
| |
|
73. Champa, D. et al. Obatoclax kills anaplastic thyroid cancer cells by inducing lysosome neutralization and necrosis. Oncotarget 7, 34453-34471 (2016). https://doi.org/10.18632/oncotarget.9121 PMid:27144341 PMCid:PMC5085168
|
|
| |
|
74. Hong, X.-Q., He, X.-Y., Tam, K. Y. & Chen, W.-H. Synthesis and biological effect of lysosome-targeting fluorescent anion transporters with enhanced anionophoric activity. Bioorg. Med. Chem. Lett. 30, 127461 (2020). https://doi.org/10.1016/j.bmcl.2020.127461 PMid:32755679
|
|
| |
|
75. Zasadny;, F. M., Ankrum;, J. A. & Abel, E. D. Lysosomal reacidification via degradation of PLGA nanoparticles in a lipotoxic cardiomyopathy model. in (Front. Bioeng. Biotechnol. Conference Abstract: 10th World Biomaterials Congress., 2016). doi:10.3389/conf.FBIOE.2016.01.01307. https://doi.org/10.3389/conf.FBIOE.2016.01.01307
|
|
| |
|
76. Zeng, J., Shirihai, O. S. & Grinstaff, M. W. Degradable Nanoparticles Restore Lysosomal pH and Autophagic Flux in Lipotoxic Pancreatic Beta Cells. Adv. Healthc. Mater. 8, (2019). https://doi.org/10.1002/adhm.201801511 PMid:30698920
|
|
|