Background
Cardiovascular disease (CVD), the leading cause of death and disability in developed countries, arises primarily from loss of or damage to populations of cardiomyocytes (CMs). Much effort has been invested to explore strategies that could promote cardiac repair, including injection of human CMs derived from embryonic stem cells (hESCs) or induced pluripotent stem cells (iPSCs) into the heart, reprogramming of non-myocardial cells into CMs, and stimulation of CM proliferation. However, success with these applications remains limited by several obstacles including unstable differentiation efficiency, immaturity and poor survival after transplantation. Therefore, understanding mechanisms underlying normal cardiac development and initiation of CVD is important to develop novel therapeutic strategies for heart disease and regenerative medicine.
Cardiac development is exquisitely sensitive to the precise temporal regulation of thousands of genes that govern developmental decisions during lineage commitment, and disruption of transcriptional networks underlies abnormal cardiac development as well as CVD(1) (2). Recently, several studies demonstrated that coordinated transition of epigenetic modifications plays a critical role in stage-specific gene expression during cardiac differentiation, with relevance to developmental birth defects including congenital cardiomyopathies (2) (4). The epigenetic landscape of the cardiac genome changes dramatically during CVD, demonstrating that epigenetic regulation also plays a previously unappreciated role in the pathogenesis of cardiac disease in adults (5) (6) . This review summarizes our current understanding of epigenetic control mechanisms regulating cardiac development and cardiovascular diseases, particularly focuses on the function of DNA methylation and demethylation through families of DNA methyltransferases and dioxygenases.
Epigenetic Regulation by DNA Methylation and Demethylation
In vertebrates, the most common DNA methylation event produces 5-methylcytosine (5mC) at CpG dinucleotide sites, which is important for proper gene expression, splicing and genome stability. DNA methylation, especially in promoter regions, is usually negatively correlated with gene activation(7). DNA methyltransferases (DNMTs) are enzymes that catalyze the transfer of methyl groups typically from the S-Adenosyl methionine donor substrate onto cytosine to yield 5mC. In mammals, DNMT1 plays a role in maintaining DNA methylation during replication, while new DNA methylation patterns are established by de novo DNA methyltransferases, DNMT3A and 3B. Depletion of DNMT1 in hESCs initiates rapid, global loss of DNA methylation, followed by extensive cell death. Upon loss of DNMT3A and DNMT3B, hESCs retain pluripotency with distinct effects on the DNA methylation landscape (8). Deletion of DNMTs in mice results in embryonic (Dnmt1, Dnmt3b) or postnatal (Dnmt3a) lethality, confirming their essential roles in development (9-10). Methylation of cytosine at non-CpG sites is rare in most mammalian cell types, but recent studies suggest non-CpG methylation is relatively abundant in embryonic stem cells and oocytes (11-12).
DNA demethylation can take place either passively or actively. Passive loss of 5mC can be achieved through DNA replication in the absence of functional DNMT1, which plays an important role in the global erasure of 5mC in the maternal genome during mouse preimplantation development and in the transition of human embryonic stem cells from primed to naïve state (13-14). 5mC can also be rapidly erased independently of DNA replication. In mammals, the ten-eleven translocation (TET) DNA dioxygenases (TET1, TET2, and TET3) were identified as the enzymes that convert 5mC to 5-hydroxymethylcytosine (5hmC), and subsequently to 5-formylcytosine (5fC) and 5-carboxycytosine (5caC), to facilitate active DNA demethylation in specific biological settings (figure 1) (15-16). Depletion of all three TET genes in hESCs showed a complete loss of 5hmC yet the cells retained pluripotency. However, they failed to form teratomas, suggesting a particularly important role for differentiation (17). In mice, inactivation of all three Tet genes leads to gastrulation phenotypes, including primitive streak patterning defects and impaired mesoderm development. TETs modulate Lefty-Nodal signaling by promoting demethylation in opposition to DNMT3A/B-mediated DNA methylation, demonstrating that dynamic DNA methylation and demethylation is critical in early body plan formation (18).
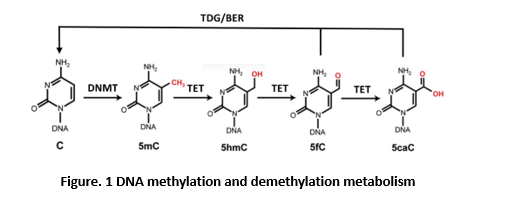
The step-wise cytosine modification pathway whereby DNMTs methylate 5C resulting in 5mC, which can be further modified by TET enzymes to 5hmC, 5fC, and 5caC. Enzymes of the TDG/BER pathway further excise and remove oxidized 5mC bases (5fC/5caC) to complete DNA demethylation. TDG, thymine-DNA glycosylase; BER, base-excision repair.
DNA Methylation and Cardiac Development
The heart is the first organ to form during mammalian embryogenesis. In mice, cardiogenesis begins around embryonic day (E) 6.5 shortly after gastrulation. Embryonic cardiomyocytes form from two embryonic fields: the first heart field gives rise to the left ventricle and some atrial tissue, while the second heart field gives rise to the right ventricle, outflow tract, interventricular septum and also contributes atria. Derived from a common early progenitor, non-myocardial cells including endothelial and smooth muscle cells, add to the myocardial lineages to form the heart (1-3). Several studies have associated DNA methylation with cardiac development using both animal model and in vitro culture systems. DNA methylation profiling of developing mouse embryonic hearts found that global methylation remains relatively stable between E11.5 and E14.5. However, differential methylation occurs at a small fraction of genes highly associated with cardiac development and revealing a regulatory relationship between differential DNA methylation and cardiac gene expression, including Has2, a gene required for heart valve formation (19). In addition, genome-wide DNA methylation analysis comparing hESCs and hESCs-derived CMs showed that CMs display higher global DNA methylation levels compared with hESCs, Taken together, the dynamics of mCpG and gene expression indicates an important regulatory role of DNA methylation during cardiac development and maturation. Additional evidence comes from studies that manipulate DNA methylation modifying enzymes. Knockdown of Dnmt3a causes disrupted sarcomere assembly, decreased beating frequency, and defective contractile movement in mouse embryonic CMs. This is associated with decreased methylation levels in the promoters of Myh7, Myh7b, Tnni3, and Tnnt2, consistent with the up-regulation of these genes caused by DNMT3A siRNA (23). Similarly, deletion of Dnmt3a and Dnmt3b genes prevents postnatal de novo methylation of fetal troponin I isoform (Tnni1) and partially relieves repression of Tnni1 (24). These data are consistent with DNA methylation profiling of embryonic and postnatal CMs (21), suggesting that de novo methylation of embryonic developmental genes by DNMT3A/B is important for the perinatal switch in sarcomere protein isoforms and postnatal cardiomyocyte maturation. Compared with DNMTs, the functions of TET enzymes during cardiac development are less studied. Tet3-deficient mouse embryonic stem cells showed impaired neural conversion, with skewing toward cardiac mesoderm by modifying Wnt signaling (25). In the zebrafish model, tet2/3 regulate proepicardial cell migration by regulating genes associated with extracellular matrix (ECM) organization. Larvae mutant for tet2 and tet3 fail to demethylate genes encoding Inhbaa in endocardium and Sox9b in myocardium, leading to defects in extracellular matrix needed to form valves and to recruit epicardial progenitors onto the heart tube (26). However, how active DNA nuclei suggested that disease-associated changes in gene expression are accompanied by concordant alterations in active histone marks, while disease-associated alterations of mCpG are rare and not directly linked to gene expression (21), which is conflict with other studies (29-30). One possible reason for the conflict is that previous studies were performed in heart tissue rather than purified CMs and therefore identified disease-associated DMRs (differentially methylated regions) could potentially arise from genes in non-cardiomyocyte lineages in the heart. Another complication is that bisulfite sequencing, which is used to resolve DNA methylation patterns at the single nucleotide level, does not discriminate between 5mC and 5hmC. A study using CMs from failing murine hearts showed also that mCpG is relatively stable in HF (24). However, using less stringent criteria, they identified more disease-associated DMRs, and demonstrated that methylation levels at these DMRs partially resemble the newborn CpG methylation pattern. Importantly, the 5hmC landscape, representing an intermediate in TET-mediated DNA demethylation, shows a similar pattern shift. Recent studies demonstrated that 5hmC is also an epigenetic feature that accompanies transcriptional changes (31). 5hmC marks the body of highly expressed genes as well as distal regulatory regions with enhancer activity. Further analysis demonstrated that 5hmC levels decrease during the induction of cardiac hypertrophy. Genome-wide distribution of 5hmC positively correlates with gene expression and shifts toward a neonatal like distribution pattern following pathological hypertrophy (31). Mechanistically, absolute 5hmC content correlated positively with Tet2 expression. modification, ATP-dependent chromatin remodeling and non-coding RNAs co-occur and are closely inter-connected during cardiac development and CVD. Novel technologies with integrative approaches make it feasible to combine different layers of epigenetic landscapes in a spatiotemporal manner and further address how the interplay between different epigenetic mechanisms relates to cardiac development and disease. These studies will be invaluable
DNA methylation profiling was also performed comparing fetal, infant and adult human CMs to study the cardiac epigenome during prenatal development and postnatal maturation. From fetal to adult stages, loss of mCpG at cis-regulatory and genic regions is associated with increased expression of genes related with myofibril or sarcomere structures as well as contraction. In contrast, a developmental increase of mCpG is linked to decreased gene expression of primarily developmental genes, including genes related to stem cell proliferation (MYC, SOX11) and cardiac ventricle morphogenesis (TNNI1, TBX5, SOX11)(21). Notably, similar to what was found during hESC to embryonic CM differentiation, a large group of early developmental genes including most cardiac TFs maintain low mCpG despite transcriptional activity changed during development, indicating their expression is largely independent of DNA methylation. Moreover, an array-based DNA methylation analysis defined a subset of 16 differentially methylated CpG loci enabling characterization of human atrial and ventricular cardiac tissue samples (22). demethylation is targeted to regulate specific genes during cardiac development remains largely unknown. Since during prenatal development and postnatal maturation, regions that lose mCpG are pre-marked with 5hmC (21), TETs might play functions in activating these genes through active DNA demethylation.
DNA Methylation and CVD
While DNA methylation was originally identified playing roles in the establishment of cellular identity during embryogenesis, a number of recent studies suggest that changes in DNA methylation states is also associated with adult diseases, including CVD pathophysiology. Cardiovascular risk factors such as smoking and high fat diet result in dysregulated DNA methylation (27). DNA methylation-based estimation of “biology age” is well correlated with incident of cardiovascular disease risk in a 10-year follow-up study (28). Moreover, several studies have identified altered mCpG patterns in the hearts of patients suffering from dilated cardiomyopathy (DCM), ischemic cardiomyopathy (ICM) and heart failure (HF) (5) (21) (29-30). Combining DNA methylation patterns with gene expression analyses, several new candidate genes not previously linked with CVD were identified with altered methylation status correlated with abnormal gene expression, including DUX4 in end-stage HF, and LY75 and ADORA2a in DCM (29-30). Knockdown of these genes in zebrafish induces cardiac dysfunction, indicating these genes play functions relevant to CVD pathophysiology. However, compared with developmental dynamics, the observed changes of CpG methylation in CVD is much smaller in scale. A genome-wide methylation study using human adult failing and non-failing CM Depletion of 5hmC via knockdown down of Tet2 affects the expression of key cardiovascular genes, including Myh7, Mycod, Srf, and Klf4 (31). In addition to TET enzymes, DNMT3A is downregulated in patients with tetralogy of Fallot (TOF) and upregulated in activated cardiac fibroblasts, leading to cardiac fibrosis (32-33). Dnmt3a heterozygous KO mice have reduced heart rate variability, which may induce adult heart failure (34). These data demonstrate both TETs and DNMTs play important roles during CVD pathophysiology
Conclusions and Perspectives
Knowledge of the role for DNA methylation modification in cardiac development and CVD has substantially increased over the last decade. It has become clear that DNA methylation and demethylation contribute to cardiac embryonic development, postnatal maturation, as well as CVD pathophysiology. However, our understanding of DNA methylation regulation during cardiovascular development and disease remains limited. It is challenging to determine if DNA methylation modification and transcriptional output are causally related, or if methylation changes are merely associated with another regulatory mechanism. In some cases it is still debatable whether DNA methylation changes in CVD are broad or negligible. Additional studies need to investigate more thoroughly whether observed DNA methylation changes in fact translate to downstream biological effects. Although the scope of this review focuses on DNA methylation and demethylation, it should be noted that multiple epigenetic regulatory mechanisms such as histone in understanding the molecular mechanisms involved in cardiac development and disease and serve as a roadmap for hESC or iPSC based therapies to regenerate and repair injured cardiac tissues.
Acknowledgement:
The authors gratefully acknowledge support from the National Institutes of Health (R35 HL135778 to T.E.).
References
1. Evans S, Yelon D, Conlon F, Kirby M. Myocardial Lineage Development. Circulation Research. 2010;107(12):1428-1444.https://doi.org/10.1161/CIRCRESAHA.110.227405 PMid:21148449 PMCid:PMC3073310
2. Wamstad J, Alexander J, Truty R, Shrikumar A, Li F, Eilertson K et al. Dynamic and Coordinated Epigenetic Regulation of Developmental Transitions in the Cardiac Lineage. Cell. 2012;151(1):206-220. https://doi.org/10.1016/j.cell.2012.07.035 PMid:22981692 PMCid:PMC3462286
3. Burridge P, Sharma A, Wu J. Genetic and Epigenetic Regulation of Human Cardiac Reprogramming and Differentiation in Regenerative Medicine. Annual Review of Genetics. 2015;49(1):461-484. https://doi.org/10.1146/annurev-genet-112414-054911 PMid:26631515 PMCid:PMC4962619
4. Lee J, Shao N, Paik D, Wu H, Guo H, Termglinchan V et al. SETD7 Drives Cardiac Lineage Commitment through Stage-Specific Transcriptional Activation. Cell Stem Cell. 2018;22(3):428-444.e5. https://doi.org/10.1016/j.stem.2018.02.005 PMid:29499155 PMCid:PMC5929163 embryonic stem cells. Nature Genetics. 2015;47(5):469-478. https://doi.org/10.1038/ng.3258 PMid:25822089 PMCid:PMC4414868
5. Pepin M, Ha C, Crossman D, Litovsky S, Varambally S, Barchue J et al. Genome-wide DNA methylation encodes cardiac transcriptional reprogramming in human ischemic heart failure. Laboratory Investigation. 2018;99(3):371-386. https://doi.org/10.1038/s41374-018-0104-x PMid:30089854 PMCid:PMC6515060
6. Meder B, Haas J, Sedaghat-Hamedani F, Kayvanpour E, Frese K, Lai A et al. Epigenome-Wide Association Study Identifies Cardiac Gene Patterning and a Novel Class of Biomarkers for Heart Failure. Circulation. 2017;136(16):1528-1544. https://doi.org/10.1161/CIRCULATIONAHA.117.027355 PMid:28838933
7. Smith Z, Meissner A. DNA methylation: roles in mammalian development. Nature Reviews Genetics. 2013;14(3):204-220. https://doi.org/10.1038/nrg3354 PMid:23400093
8. Liao J, Karnik R, Gu H, Ziller M, Clement K, Tsankov A et al. Targeted disruption of DNMT1, DNMT3A and DNMT3B in human embryonic stem cells. Nature Genetics. 2015;47(5):469-478. https://doi.org/10.1038/ng.3258 PMid:25822089 PMCid:PMC4414868
9. Okano M, Bell D, Haber D, Li E. DNA Methyltransferases Dnmt3a and Dnmt3b Are Essential for De Novo Methylation and Mammalian Development. Cell. 1999;99(3):247-257. https://doi.org/10.1016/S0092-8674(00)81656-6
10. Li E, Bestor T, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69(6):915-926. https://doi.org/10.1016/0092-8674(92)90611-F
11. Shirane K, Toh H, Kobayashi H, Miura F, Chiba H, Ito T et al. Mouse Oocyte Methylomes at Base Resolution Reveal Genome-Wide Accumulation of Non-CpG Methylation and Role of DNA Methyltransferases. PLoS Genetics. 2013;9(4):e1003439. https://doi.org/10.1371/journal.pgen.1003439 PMid:23637617 PMCid:PMC3630097
12. Xie W, Barr C, Kim A, Yue F, Lee A, Eubanks J et al. Base-Resolution Analyses of Sequence and Parent-of-Origin Dependent DNA Methylation in the Mouse Genome. Cell. 2012;148(4):816-831. https://doi.org/10.1016/j.cell.2011.12.035 PMid:22341451 PMCid:PMC3343639
13. Guo H, Zhu P, Yan L, Li R, Hu B, Lian Y et al. The DNA methylation landscape of human early embryos. Nature. 2014;511(7511):606-610. https://doi.org/10.1038/nature13544
14. von Meyenn F, Iurlaro M, Habibi E, Liu N, Salehzadeh-Yazdi A, Santos F et al. Impairment of DNA Methylation Maintenance Is the Main Cause of Global Demethylation in Naive Embryonic Stem Cells. Molecular Cell. 2016;62(6):848-861. https://doi.org/10.1016/j.molcel.2016.04.025 PMid:27237052 PMCid:PMC4914828
15. He Y, Li B, Li Z, Liu P, Wang Y, Tang Q et al. Tet-Mediated Formation of 5-Carboxylcytosine and Its Excision by TDG in Mammalian DNA. Science. 2011;333(6047):1303-1307. https://doi.org/10.1126/science.1210944 PMid:21817016 PMCid:PMC3462231
16. Ito S, Shen L, Dai Q, Wu S, Collins L, Swenberg J et al. Tet Proteins Can Convert 5-Methylcytosine to 5-Formylcytosine and 5-Carboxylcytosine. Science. 2011;333(6047):1300-1303. https://doi.org/10.1126/science.1210597 PMid:21778364 PMCid:PMC3495246
17. Verma N, Pan H, Doré L, Shukla A, Li Q, Pelham-Webb B et al. TET proteins safeguard bivalent promoters from de novo methylation in human embryonic stem cells. Nature Genetics. 2017;50(1):83-95. https://doi.org/10.1038/s41588-017-0002-y PMid:29203910 PMCid:PMC5742051
18. Dai H, Wang B, Yang L, Chen J, Zhu G, Sun M et al. TET-mediated DNA demethylation controls gastrulation by regulating Lefty-Nodal signalling. Nature. 2016;538(7626):528-532. https://doi.org/10.1038/nature20095 PMid:27760115
19. Chamberlain A, Lin M, Lister R, Maslov A, Wang Y, Suzuki M et al. DNA Methylation is Developmentally Regulated for Genes Essential for Cardiogenesis. Journal of the American Heart Association. 2014;3(3). https://doi.org/10.1161/JAHA.114.000976
20. Gu Y, Liu G, Plongthongkum N, Benner C, Yi F, Qu J et al. Global DNA methylation and transcriptional analyses of human ESC-derived cardiomyocytes. Protein & Cell. 2014, 5(1):59-68 https://doi.org/10.1007/s13238-013-0016-x PMid:24474197 PMCid:PMC3938846
21. Gilsbach R, Schwaderer M, Preissl S, Grüning B, Kranzhöfer D, Schneider P et al. Distinct epigenetic programs regulate cardiac myocyte development and disease in the human heart in vivo. Nature Communications. 2018;9(1). https://doi.org/10.1038/s41467-017-02762-z PMid:29374152 PMCid:PMC5786002
22. Hoff K, Lemme M, Kahlert A, Runde K, Audain E, Schuster D et al. DNA methylation profiling allows for characterization of atrial and ventricular cardiac tissues and hiPSC-CMs. Clinical Epigenetics. 2019;11(1). https://doi.org/10.1186/s13148-019-0679-0 PMid:31186048 PMCid:PMC6560887
23. Fang X, Poulsen R, Wang-Hu J, Shi O, Calvo N, Simmons C et al. Knockdown of DNA methyltransferase 3a alters gene expression and inhibits function of embryonic cardiomyocytes. The FASEB Journal. 2016;30(9):3238-3255. https://doi.org/10.1096/fj.201600346R PMid:29231988
24. Gilsbach R, Preissl S, Grüning B, Schnick T, Burger L, Benes V et al. Dynamic DNA methylation orchestrates cardiomyocyte development, maturation and disease. Nature Communications. 2014;5(1). https://doi.org/10.1038/ncomms6288 PMid:25335909 PMCid:PMC4220495
25. Li X, Yue X, Pastor W, Lin L, Georges R, Chavez L et al. Tet proteins influence the balance between neuroectodermal and mesodermal fate choice by inhibiting Wnt signaling. Proceedings of the National Academy of Sciences. 2016;113(51):E8267-E8276. https://doi.org/10.1073/pnas.1617802113 PMid:27930333 PMCid:PMC5187696
26. Lan Y, Pan H, Li C, Banks K, Sam J, Ding B et al. TETs Regulate Proepicardial Cell Migration through Extracellular Matrix Organization during Zebrafish Cardiogenesis. Cell Reports. 2019;26(3):720-732.e4. https://doi.org/10.1016/j.celrep.2018.12.076 PMid:30650362 PMCid:PMC6366638
27. Zhong J, Agha G, Baccarelli A. The Role of DNA Methylation in Cardiovascular Risk and Disease. Circulation Research. 2016;118(1):119-131. https://doi.org/10.1161/CIRCRESAHA.115.305206 PMid:26837743 PMCid:PMC4743554
28. Lind L, Ingelsson E, Sundström J, Siegbahn A, Lampa E. Methylation-based estimated biological age and cardiovascular disease. European Journal of Clinical Investigation. 2017;48(2):e12872. https://doi.org/10.1111/eci.12872 PMid:29231988
29. Movassagh M, Choy M, Knowles D, Cordeddu L, Haider S, Down T et al. Distinct Epigenomic Features in End-Stage Failing Human Hearts. Circulation. 2011;124(22):2411-2422. https://doi.org/10.1161/CIRCULATIONAHA.111.040071 PMid:22025602 PMCid:PMC3634158
30. Haas J, Frese KS, Park YJ, Keller A, Vogel B, Lindroth AM et al (2013) Alterations in cardiac DNA methylation in human dilated cardiomyopathy. EMBO Mol Med 5:413-429https://doi.org/10.1002/emmm.201201553 PMid:23341106 PMCid:PMC3598081
31. Greco C, Kunderfranco P, Rubino M, Larcher V, Carullo P, Anselmo A et al. DNA hydroxymethylation controls cardiomyocyte gene expression in development and hypertrophy. Nature Communications. 2016;7(1). https://doi.org/10.1038/ncomms12418 PMid:27489048 PMCid:PMC4976219
32. SHENG W, QIAN Y, WANG H, MA X, ZHANG P, CHEN L et al. Association between mRNA levels of DNMT1, DNMT3A, DNMT3B, MBD2 and LINE-1 methylation status in infants with tetralogy of Fallot. International Journal of Molecular Medicine. 2013;32(3):694-702. https://doi.org/10.3892/ijmm.2013.1427 PMid:23820632
33. Tao H, Yang J, Chen Z, Xu S, Zhou X, Zhan H et al. DNMT3A silencing RASSF1A promotes cardiac fibrosis through upregulation of ERK1/2. Toxicology. 2014;323:42-50. https://doi.org/10.1016/j.tox.2014.06.006 PMid:24945829
34. Ponikowski P, Anker S, Chua T, Szelemej R, Piepoli M, Adamopoulos S et al. Depressed Heart Rate Variability as an Independent Predictor of Death in Chronic Congestive Heart Failure Secondary to Ischemic or Idiopathic Dilated Cardiomyopathy. The American Journal of Cardiology. 1997;79(12):1645-1650. https://doi.org/10.1016/S0002-9149(97)00215-4