Abstract
The circadian clock regulates vital cellular processes by adjusting the physiology of the organism to daily changes in the environment. Rhythmic transcription of core Clock Genes (CGs) and their targets regulate these processes at the cellular level. Circadian clock disruption has been observed in people with neurodegenerative disorders like Alzheimer’s and Parkinson’s. Also, ablation of CGs during development has been shown to affect neurogenesis in both in vivo and in vitro models. Previous studies on the function of CGs in the brain have used knock-out models of a few CGs. However, a complete catalogue of CGs in different cell types of the developing brain is not available and it is also tedious to obtain. Recent advancements in single-cell RNA sequencing (scRNA-seq) have revealed novel cell types and elusive dynamic cell states of the developing brain. In this study, by using publicly available single-cell transcriptome datasets we systematically explored CGs co-expressing networks (CGs-CNs) during embryonic and adult neurogenesis. Our meta-analysis reveals CGs-CNs in human embryonic radial glia, neurons, and other lesser studied non-neuronal cell types of the developing brain.
Keywords: Circadian clock; neurogenesis; single-cell transcriptome; SCENIC; TSCAN.
Introduction
Self -sustained oscillatory expression of a core set of genes is known to drive circadian rhythms in organisms ranging from fungi and archaebacteria to humans. At the molecular level, these rhythms arise from the waxing and waning of transcription factors which gives rise to rhythmicity in gene expression of their targets. In recent years, several groups have shown the association of key components of the molecular clock with neuronal function and diseases. Specifically, CLOCK protein, a part of the positive arm of the transcription-translation feedback loop has been shown to affect migration of neuronal cells(1,2,3). The reduced expression of CLOCK has been observed in epileptogenic tissues in humans(4). Also, knock-out of BMAL1 the binding partner of CLOCK, in mice has been shown to lower the seizure threshold(5). Circadian disruption has also been observed in major depressive disorder (MDD), as the rhythmicity of CGs like BMAL1, PER1-2-3, NR1D1, BHLHE40-41, and DBP was disrupted in the post-mortem brains derived from MDD patients(6).
The development and function of the brain are maintained in large part by embryonic and adult neurogenesis. During cortical development, heterogeneous populations of neural stem cells are present in the subventricular zone (SVZ) and ventricular zone while differentiating neurons form cortical plate. Previous studies have shown the involvement of CGs in neurogenesis; BMAL1 and PER2 are involved in cell cycle regulation in adult neurogenesis(7). Also, BMAL1 knockout mice exhibit symptoms of aging and perturbed ocular parameters(8). Similarly, conditional mutant mice of Clock or Bmal1 show delayed maturation of inhibitory parvalbumin neurons in the visual cortex(9). The above studies clearly establish the role of CGs in brain development, through genetic manipulation of selected CGs. Systematically extending the above strategies to all CGs is both laborious and time consuming. In the case of the developing human brain, it is further complicated due to ethical reasons. In this study, we analysed scRNA-seq datasets of embryonic and adult neurogenic niches to comprehensively explore the cell type identity
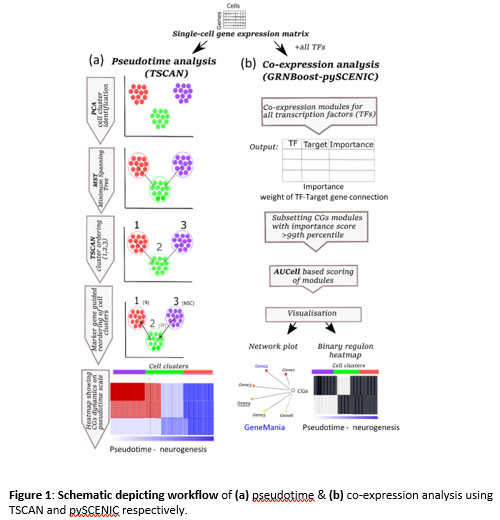
Figure 1: Schematic depicting workflow of (a) pseudotime & (b) co-expression analysis using TSCAN and pySCENIC respectively.
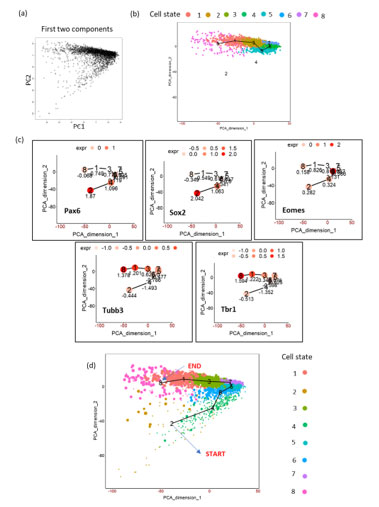
FigureS1. Schematics of TSCAN workflow on mouse embryonic cortex (E15.5) (a) PCA plot on the dataset showing first two PCs. (b) TSCAN MST and its cell state ordering (8,1,2,6,5,7). Cell state 2 and 4 were omitted by TSCAN. (c) Expression of markers genes by manually including all cell states. (d) Marker gene expression based manual re-ordering of pseudotime (2,4,6,5,7,3,1,8).
and expression dynamics of CGs in neurogenesis using pseudotime analysis. Since most of the CGs function as transcription factors (TFs) with a wide range of genomic targets(10), we also identified their co-expressing networks (CNs). For pseudotime and co-expression analysis, we used TSCAN (pseudo-Time reconstruction in Single-Cell RNA-seq Analysis)(11) and SCENIC (Single-Cell rEgulatory Network Inference and Clustering)(12) respectively, for more description refer to methods section (Figure1, S1). Our analysis reveals CGs-CNs enrichment in distinct cell types of the neurogenic niches thereby providing a framework for future studies exploring clock genes in brain development.
Results
For the list of CGs that form the focus of this study refer to supplementary TableS1. We followed the recommendation of HUGO Gene Nomenclature Committee for gene names. Hence gene names are in upper case (e.g., CLOCK) for humans and the first letter in upper case for mouse (e.g., Clock).
Expression dynamics of CGs-CNs in mouse embryonic cortex E15.5
Embryonic neurogenesis is characterized by the rapid proliferation of neural stem cells. These cells divide symmetrically and expand the pool of neurogenic stem cells during the initial stages of neurogenesis. Later stages of embryonic neurogenesis are characterized by asymmetric divisions to give rise to neurons via intermediate progenitor cells(13). In contrast to embryonic neurogenesis, adult neurogenesis is restricted to specialized regions like SVZ and subgranular zone (SGZ)(14). Apart from giving rise to neurons and glia, embryonic neural stem cells also serve as the source of adult neural stem cells. We intended to analyze the expression of CGs with cell type identity and expression dynamics in mouse embryonic and adult neurogenesis. To study the CGs expression dynamics during embryonic neurogenesis we have used a single-cell transcriptome dataset of mice generated by Yuzwa et. al., which encompass embryonic timepoints: E11.5, E13.5, E15.5 and E17.5 days(15). Using (TSCAN), neural progenitors and differentiating neuron clusters were identified from the above datasets. Pax6 and Tubb3 marker genes were used to identify neural progenitors and differentiating neurons, respectively. Further, these cells were pseudotime aligned with neural progenitors at the start and differentiating neurons at the end. Upon this neurogenesis trajectory aligned by pseudotime, the statistical significance of expression dynamics of CGs was calculated. We applied TSCAN workflow to all the developmental time points (data not shown), only E15.5 showed statistically significant expression dynamics for three CGs – Rorb, Nfil3 and Csnk1e (Figure 2a). Except for Csnk1e, the two CGs expressions were high along the pseudotime associated with differentiated neurons. The lack of expression dynamics of CGs in other three times points (E11.5, E13.5 and E17.5) may be due to high drop-out rates observed in scRNA-Seq or true CGs expression trend, addressing the above issue is beyond the scope of this study.
Applying the co-expression analysis workflow on the E15.5 dataset revealed two gene modules: Hlf(21g) and Rorb(51g) (Figure 2b). Hitzler et al. earlier hypothesized the role of Hlf in neuronal differentiation based on its expression pattern in diverse regions of developing mouse brain(16), and also experimental models of epilepsy show downregulation of Hlf(17). Rorb shows expression in the L4 region of the mouse and human brain(18). It also plays a vital role in the development and organization of the whisker barrel in rodent brain(19). These studies provide hints for the involvement of Hlf and Rorb in brain development. Additionally, our analysis discloses their underlying gene regulatory network (Figure 2c & S3). Out of 21 genes in the Hlf module (Figure 2c), Ntrk2 was previously reported in the GeneMANIA co-expression database.
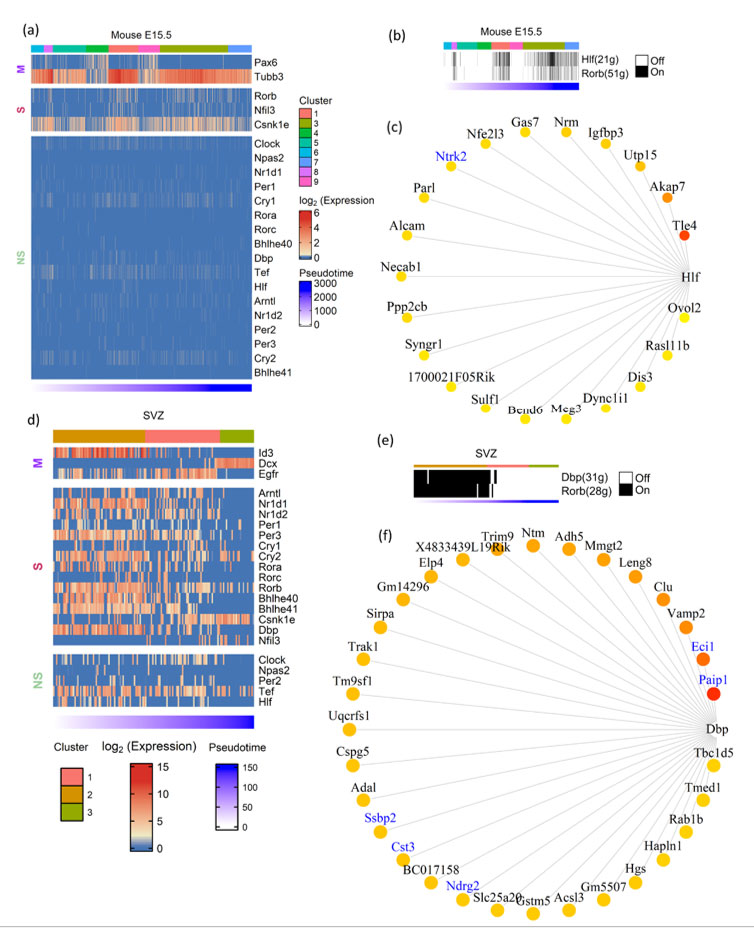
Figure 2: Expression dynamics of CGs-CNs in mouse embryonic and adult neurogenesis. a&d) Heatmap showing CGs (row) expression over pseudotime ordered cells (column) in neurogenesis trajectory of (a) mouse E15.5 cortex, (d) mouse SVZ), top annotation denotes cell clusters assigned by TSCAN, bottom annotation denotes TSCAN calculated pseudotime in ascending order. In heatmaps, M denoting marker genes, S and NS denoting statistically significant and non-significant pseudotime expression dynamics, respectively. (b&e) Binary activity heatmap of CG modules over pseudotime aligned neurogenesis trajectory of (b) mouse E15.5 cortex (e) mouse SVZ. Top annotation denotes cell clusters assigned by TSCAN, bottom annotation denotes TSCAN calculated pseudotime in ascending order. c&f) Network plot showing CGs and its target genes, the edges of the target genes placed from highest importance score (red) to lowest (yellow). GeneMANIA reported co-expression gene pairs are indicated in blue color.
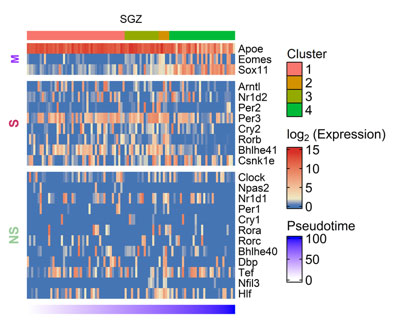
Figure S2. Heatmap showing CGs (row) expression over pseudotime ordered cells (column) in mouse SGZ, M denoting marker genes, S and NS depicting genes showing statistically significant and non-significant pseudotime analysis.
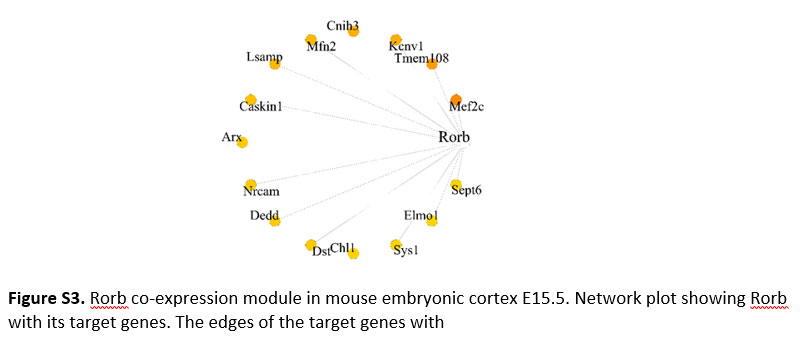
Figure S3. Rorb co-expression module in mouse embryonic cortex E15.5. Network plot showing Rorb with its target genes. The edges of the target genes with
Expression dynamics of CGs-CNs in the neurogenic niches of adult mouse brain
In the adult brain, neurogenesis occurs in restricted niches: SGZ of the hippocampus and SVZ near the lateral ventricles(20,21). The periodic activation of quiescent neural stem cells (qNSCs) in SGZ/SVZ produces mature neurons migrating to deep layers of dentate gyrus and olfactory bulb respectively, whose homeostasis plays a vital role in learning, memory formation and emotion regulation(22,23). SGZ and SVZ regions are shown to have expressions of CGs(7). Using publicly available scRNA-seq data and pseudotime analysis, we studied the CGs expression dynamics during adult neurogenesis.
For pseudotime ordering of SVZ(24) we used known marker genes like Id3 and Egfr for quiescent and activated NSCs respectively and Dcx for differentiated neurons (Figure 2d). Most of the CGs analyzed showed significant enrichment in the initial stages of pseudotime which corresponds to the transition from quiescent to activated neural stem cells. Though most of the CGs analyzed showed enrichment during the early stages of adult neurogenesis, Csnk1e showed reverse trend of enrichment, with higher expression in the later stages of pseudotime corresponding to the birth of differentiated neurons.
Besides SVZ, we also analyzed SGZ, the other prominent adult neurogenic niche using scRNA-seq data(25). It also showed enrichment of CGs expression during the early stages of neurogenesis (Figure S2), though the expression dynamics were not as evident as in SVZ. Both SVZ and SGZ showed enrichment of Csnk1e during later stages of adult neurogenesis. Our co-expression analysis on SVZ and SGZ, revealed CGs-CNs only in SVZ datasets as none of the predicted CNs of SGZ datasets passed the AUCell scoring threshold. In SVZ, the co-expression analysis revealed two distinct CGs-CNs: Dbp(31g) and Rorb(28g) active during early stages of adult neurogenesis (Figure 2e). The detailed analysis of the Dbp module (Figure 2f) revealed 5 of its target genes (Paip1, Eci1, Ssbp2, Cst3 and Ndrg2) to be documented in the GeneMANIA. Among the above genes, Ndrg2 expression was previously reported in SVZ(26).
Expression dynamics of CGs-CNs in developing human brain
The development of the human brain and maturation lasts upto to 20 years(27), while embryonic and fetal developmental changes range from 0-8 and 8-24 post-conception week (pcw) respectively(28). To identify CGs-CNs in early human brain development, we analyzed the scRNA-seq dataset generated by Nowakowski et al.(29). The dataset includes prefrontal cortex (PFC) and primary visual cortex (V1), with ages ranging from 5 to 37pcw. In total, from 48 samples they broadly identified 11 major cell types. Their dataset is also available as an interactive browser (https://cells.ucsc.edu). We applied co-expression analysis workflow on their dataset and used the cell cluster information provided by authors (Table S3). Our co-expression analysis revealed four CGs-CNs: BHLHE41(193g), BHLHE40(57g). RORB(12g) and NR1D2(14g) (Figure 3a).
Interestingly, the BHLHE41(193g)/BHLHE40(57g) module enriched predominantly in non-neuronal cell types like mural, microglial and truncated radial glia (tRG) cluster (Figure 3a). Few topmost genes co-expressing with BHLHE41 (Figure 3c) included P2RY12, CLEC7A, ATP8B4 and FYB. P2RY12 functions as a receptor for nucleotides which are released in response to CNS(30,31) to assist chemotaxis of microglia to the site of injury. CLEC7A has been shown to be linked with beta amyloid disease progression(32). Genes including CLEC7A, P2RY12 and FYB serve as a marker of microglia and we reveal their co-expression with BHLHE41 in our analysis. In BHLHE40(57g) module (Figure 3d), S100A4 was one of the co-expressing target genes, which was also reported in GeneMANIA. It also acts as a biomarker of glioblastoma(32,33). Among the top connected genes in the same module, we observed HPGDS, which is known to localize to microglia and upregulated in Alzheimer’s(34). The RORB(12g) module (Figure 3e) showed enrichment in medial ganglionic eminence-radial glia cluster 1 (MGE-RG1) and excitatory neurons-PFC 1 (EN-PFC1) cluster, the RORB module was also captured in the developing cortex and SVZ dataset of the mouse (Figure S3). Comparison of the target genes of the RORB module in mouse and humans revealed MEF2C-RORB gene pair co-expression in mouse and human embryonic cortical neurogenesis.
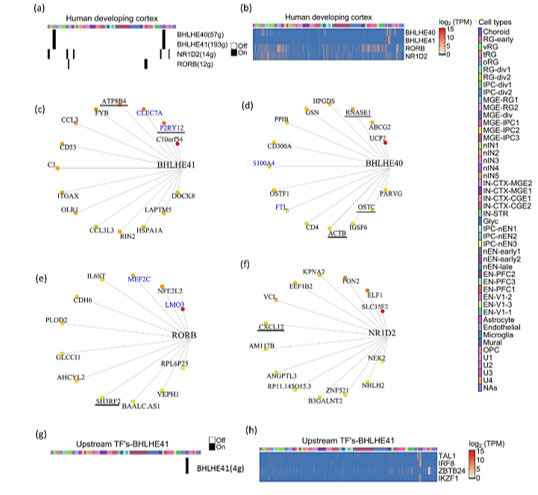
Figure3: Expression dynamics of CGs-CNs in developing human brain.
(a) Binary activity heatmap of CGs modules over cell clusters of the developing human brain, cell clusters as annotated by Nowakowski et al. for cluster description refer table S3. (b) Heatmap showing gene expression of CGs over cell clusters, which showed significant enrichment score in the co-expression analysis. Cell clusters as annotated by Nowakoski et al. for cluster description refer table S3. (c, d, e & f) Network plots showing CGs and its targets in the developing human brain, the edges are placed from highest (red) to lowest (yellow) importance score of target genes. GeneMANIA reported co-expression gene pairs are highlighted in blue color. (c) BHLHE41, (d) BHLHE40, (e) RORB and (f) NR1D2 only top15 target genes in the BHLHE40/41 module are shown, target genes with known diurnal expression are underlined. (g) Binary activity heatmap of upstream TFs co-expressing with BHLHE41 over cell clusters of the developing human brain, cell clusters as annotated by Nowakowski et al. for cluster description refer table S3. (h) Heatmap showing gene expression of upstream TFs co-expressing with BHLHE41, which showed significant enrichment score in the co-expression analysis. Cell clusters as annotated by Nowakowski et al. for cluster description refer table S3.
Knockdown of NR1D2 in adult murine neural stem cells has been shown to affect neural differentiation(37). The topmost target genes of the NR1D2 module included SLC35F2, ELF1, PON2, KPNA2 and EEF1B2 (Figure 3f). These genes are involved in vital neuronal functions like SLC35F2 in transport across blood brain barrier(38), ELF1 in neurite growth(39) and PON2 imparts neuroprotection by scavenging superoxides(38,40). The enrichment of four CGs-CNs in diverse cell types of the developing human brain prompted us to look for upstream regulators of CGs. Our search for upstream TFs co-expressing with CGs revealed only one significant upstream module associated with BHLHE41, (Figure 3g). We found four TFs namely TAL1, IRF8, IKZF1 and ZBTB24 which are documented to have a regulatory role in microglia activation in response to injury(41), aging(42) with TAL1 acting as a specific marker of human microglia(43).
Expression dynamics of CGs-CNs during astrocyte genesis
Previously, considered to be the support system of neurons in the brain, glial cells are being recognized as vital players in brain function. By promoting neuronal survival and modulating synapse function these cells actively participate in neuronal signal transduction. In particular, the participation of astrocytes in circadian regulation has been recognized recently(44). The various functions of astrocytes like modulation of intracellular calcium levels or neurotransmitter release occur rhythmically. A study(45) has shown that astrocyte specific genetic deletion of Bmal1 and behavioral disruption of rhythms in mice induces astrocyte activation. This effect is augmented when Bmal1 is knocked out in both neurons and astrocytes. These studies point to an important role for astrocytes in maintaining circadian function in the brain.
We analyzed the total transcriptome of human fetal and adult brain-derived astrocytes(46) . In contrast to neurons, astrocytes showed statistically significant upregulation of CGs namely CLOCK, CRY2, PER3, RORA, RORB, HLF, NR1D1, BHLHE40 and BHLHE41 upon maturation (Figure 4a). To strengthen this observation, we analyzed single-cell transcriptomic data of astrocytes derived from cortical spheroids(47) . Similar to neuronal differentiation analysis, we first constructed pseudotime trajectory of astrocyte differentiation and classified cell clusters using cellular markers of early and late stages of astrocyte development. Overall, we found an increased expression of all CGs except RORB and RORC in differentiated astrocytes (Figure 4b). This was in concordance with the trend seen in total RNA-seq of fetal and adult brain-derived astrocytes.
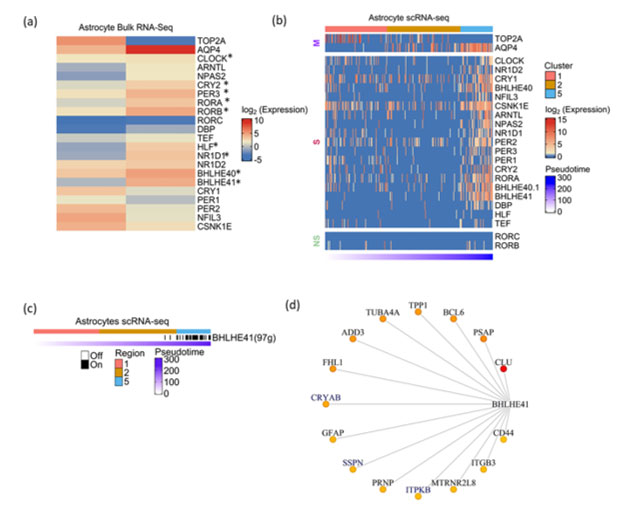
Figure 4: Expression dynamics of CGs-CNs during astrocyte genesis
a) Heatmap showing CGs expression in fetal and adult brain derived astrocytes profiled by total RNA-seq. TOP2A and AQP4 represent marker genes associated with early and late stages of astrocyte development, respectively. Asterisks indicate the differential expression with statistical significance calculated using DEseq2.b) Heatmap showing CGs (row) expression over pseudotime ordered cells (column) derived from astrocytic spheroid, M denoting marker genes, S and NS depicting genes showing statistically significant and non-significant pseudotime expression dynamics respectively. Top annotation denotes cell clusters assigned by TSCAN, bottom annotation denotes TSCAN calculated pseudotime in ascending order.
c) Binary activity heatmap of CGs modules over pseudotime aligned trajectory of astrocyte genesis. Top annotation denotes cell clusters assigned by TSCAN, bottom annotation denotes TSCAN calculated pseudotime in ascending order. d) Network plot of BHLHE41 module in astrocytes, the edges are placed from highest (red) to lowest (yellow) importance score of target genes. GeneMANIA reported co-expression gene pairs are highlighted in blue color, only top15 target genes are shown
The co-expression analysis of astrocyte scRNA-Seq revealed the BHLHE41 associated module with 97genes to be the prominent module that showed enrichment in the later stages of astrocyte genesis (Figure 4c). BHLHE41 acts as a transcriptional repressor in the circadian regulation of gene expression. Previous studies have reported the association of mutations in BHLHE41 with short sleep timing in both humans and experimental models(48,49). Also, it shows high abundance in astrocytes as compared to neurons(50). Few of the topmost target genes in the BHLHE41 module included CLU, PSAP, BCL6, CRYAB, TPP1, TUBA4A and ADD3 (Figure 4d). The co-expression of a few genes like ITPKB, SSPN and CRYAB have been previously reported in the GeneMANIA. Additionally, we report various novel genes, specifically GFAP, a well-known marker of astrocytes and also shown to be associated with the etiology of Alexander disease(51). We also observed CD44 co-expression with BHLHE41, CD44 acts as a vital adhesion protein during astrocyte development(52) and an increase in CD44 positive astrocytes have been shown in Alzheimer’s brain(53). Our approach highlights the potential regulation of these vital genes involved in astrocyte function by BHLHE41.
Discussion
We combined pseudotime/co-expression analysis and explored potential co-regulatory modules of CGs in the developing brain. Our workflow reveals CGs-CNs with cell type and cell state identity, a feat not possible from bulk RNA-seq based co-expression analysis. Also, high gene drop-out rates pertaining to scRNA-seq greatly impact the pseudotime analysis of low expressing genes like CGs. Therefore, we looked at co-expression modules that can accurately reflect variations in gene expression trends related to underlying biological processes (neurogenesis).
Our analysis of the murine and human embryonic brain scRNA-seq datasets reveals CGs-CNs of RORB, whose expression and role in cortical development is well studied in murine models. Further, many of the identified co-expression pairs are also reported in GeneMANIA with few of the target genes showing oscillatory expression in the adult brain(54). These validations highlight the biological relevance of the CGs-CNs captured through our analysis.
Many recent studies reveal the regulatory role of non-neuronal cell types in brain physiology. Our analysis also identified four CGs-CNs in the non-neuronal population of the developing human brain. Among the four modules, BHLHE41 strongly co-express with genes having a crucial function in microglia such as P2RY12 and CLEC7A. Further, our co-expression analysis for TFs upstream of CGs reveal BHLHE41 as a target of microglia associated TFs -TAL1, IRF8, IKZF1 and ZBTB24. In addition, astrocytes derived from cortical spheroids show high expression of the BHLHE41 module in mature astrocytes. Above results clearly favour BHLHE41 as a prioritized module for future biological validation in the context of glial biology.
Our meta-analysis uncovers CGs-CNs in rare cell types of the developing brain- radial glia, microglia, and astrocytes. Similar analysis workflow on scRNA-seq datasets on pre-sorted rare neuronal populations, with high sequencing depth in future can reveal more CGs-CNs in the brain. Further, a rapid increase in public availability of human scRNA-Seq dataset can be utilized to construct CGs-CN map of all cell types in human body. Additively, integrating emerging single cell chromatin accessibility maps with scRNA-seq will improve confidence of co-expression analysis in general. We also want to underlie that through this analysis we only prioritized CGs based on pseudotime and co-expression analysis in developing brain cells, understanding their functional consequence need experimental validations. Thus, studies involving genetic perturbation of these CGs in organoid models of human brain development is needed to fully appreciate the intricate role of circadian transcription factors in human brain development. Considering the known dysregulation of clock genes in psychiatric disorders it will also be interesting to extend similar analysis into scRNA-seq datasets derived from diseased conditions.
Methods
Pseudotime analysis
TSCAN was used for pseudotime analysis (https://github.com/zji90/TSCAN). Each single-cell gene expression matrix was subjected to TSCAN default workflow (Figure 1a), the workflow included preprocessing, dimensionality reduction followed by clustering. The genes with zero count were removed and scRNA-seq dropouts were handled by clustering genes having similar expression patterns using euclidean distance and complete linkage. The above pre-processing method was applied to all the datasets. Next, PCA was used for dimension reduction followed by clustering, yielding a minimum spanning tree. The identification of clusters was performed using marker genes. p-value and FDR (False discover rate) were calculated on the TSCAN ordered cells. TSCAN assigned cell clusters, were inspected for a panel of marker genes from the publications of the respective datasets, while a single marker gene was used for visualization purpose. If required, TSCAN cluster ordering was further manually corrected to place neural stem cell clusters as the starting point of the pseudotime. The TSCAN employs generalized additive model (GAM) for fitting the individual gene expression dynamics over pseudotime. GAM fitting was compared to null-model by assuming gene expression was constant over pseudotime. P-value was computed over the above comparison using likelihood ratio test and then converted to false discovery rate (FDR) using Benjamini-Hochberg Procedure. An FDR of <0.05, was used as a cut-off to score for genes with statistically significant expression dynamics over calculated pseudotime. The accession number of the datasets are provided in the supplementary table 2.
Co-expression analysis
Two steps in the pySCENIC (V0.10.3)(11)and SCENIC (Single-Cell rEgulatory Network Inference and Clustering)(12) pipeline were implemented in our co-expression analysis. First the co-expressing gene modules were identified using GRNBoost (https://github.com/aertslab/GRNBoost). Input for GRNBoost included a list of annotated transcription factors (https://github.com/aertslab/pySCENIC/tree/master/resources) and scRNA-seq expression matrix. In return GRNBoost outputs adjacency matrix with TFs, target genes and importance score. All the CGs (TFs) and its associated targets genes with importance scores greater than 99th percentile were subsetted as CGs-CNs.
Next AUCell (V1.11.0) was used for scoring the activity of above CGs-CNs. AUCell uses “Area under curve” to score the activity of a given geneset (CGs-CNs) within expressed genes of scRNA-seq dataset. AUCell was implemented using R (4.0.2) using default parameters. The CGs-CNs which crossed the default threshold of AUCell geneset activity parameters are shown as binary CGs-CNs activity heatmap.
ScRNA-seq dataset description
Mouse Cortex
Yuzwa et al. (GSE107122) performed drop-seq of CD1 mouse embryos at E11.5, 13.5, 15.5 and 17.5 stage. For E15.5 embryonic stage, 13 embryos were used and their brains dissected and dissociated for Drop-seq droplet collection. Following droplet collection single-cell libraries were prepared and sequenced on Illumina NextSeq500 platform.
Mouse Subventricular zone (SVZ)
Babodilla et al (GSE67833) performed microdissection of SVZ from C57BL/6 adult mice. GLAST+ Prominin1+ and PSA-NCAM+ were used as marker to sort neural stem cells and neuroblasts from SVZ region. scRNA-seq libraries were prepared using Smart-seq2 and sequenced on HiSeq2000 platform. Trimmed reads were mapped to the mouse genome (ENSEMBL Release 78) using STAR_2.4.0g followed by read quantification as FPKM.
Mouse Subgranular zone (SGZ)
Shin et al. (GSE71485) used transgenic mice expressing nuclear localized CFP under the control of nestin (Nes-CFPnuc). The CFP containing cells marking quiescent-NSCs and their progenies were sorted under a fluorescent microscope using glass pipette. The single-cell libraries were prepared by SMART protocol and sequenced by HiSeq2500 followed by read mapping and quantification to give TPM matrix.
Developing human fetal cortex
Nowakowski et al. performed stereotaxic based microdissection of brain regions like prefrontal cortex (PFC), primary visual cortex (V1) and MGE, the brain region aged from 5-37pcw. The single-cell suspension was prepared using FluidigmC1 followed by sequencing of single-cell libraries on the Illumina Hiseq2500 platform. The sequencing reads were aligned to GRCh38 and quantified to reveal transcript count as CPM.
Astrocyte spheroid
Astrocyte lineage cells were purified from cortical spheroids derived from human iPSC line by Sloan et al (GSE99951). The immunopanning method was used to isolate astrocytes using HepaCAM as an astrocyte marker. The sequencing libraries were prepared from astrocytes derived using immunopan method at 100, 130, 175 and 450 days of in vitro differentiation. Single-cell libraries were prepared using SMART-seq protocol and sequenced on NextSeq500 sequencing platform. The raw reads were aligned to hg19 human genome followed by transcript quantification as FPKM values.
Total RNA-seq of human astrocytes
Zhang et al. (GSE73721) performed RNA-sequencing of astrocytes derived from juvenile (8-18years old) and adult (21-63 years old) human brain. The astrocytes from the brain tissue were purified in culture using immunopanning. Briefly, single-cell suspension of donor tissues was subjected to dishes coated with antibodies against various cell types, Hepa-CAM was used as a marker for astrocytes. The cDNA libraries from the two samples were sequenced on illumina NextSeq sequencer as 150bp paired end reads. The RNA-seq reads were mapped using TopHat2 to hg19 as the reference genome and expression level was estimated as FPKM.
Acknowledgements The authors acknowledge Dr. Beena Pillai and Dr. Souvik Maiti for their help in writing the manuscript and discussions. We are grateful to them for allowing us to communicate the manuscript. We also acknowledge the high-performance-computing (HPC) facility of CSIR-IGIB.
Author contributionsSR and SS contributed equally. SR and SS conceptualized the work, SR and AHA performed the analysis, SR and SS prepared the manuscript.
Supplementary tables
Table S1. List of core clock genes analyzed in this study.
|
BMAL1/ARNTL
CLOCK
NPAS2
NR1D1
NR1D2
PER1
PER2
PER3
CRY1
CRY2
RORA
RORB
RORC
BHLHE40
BHLHE41
DBP
TEF
NFIL3
HLF
CSNK1E
|
Table S2. List of datasets used in the analysis.
| |
Accession number
|
RNA-seq platform
|
Single-cell platform
|
Enrichment of cell types
|
|
Mouse embryonic cortex (E15.5)
|
GSE107122
Yuzwa et al. 2017
|
NextSeq500
|
DropSeq
|
NO
|
|
Sub Ventricular Zone (SVZ)
|
GSE67833
Llorens-Bobadilla et al. 2015
|
HiSeq2000/MiSeq
|
FluidigmC1
|
FACS sorting
|
|
Sub Granular Zone (SGZ)
|
GSE71485
Shin et al. 2015
|
HiSeq2500
|
SMART-seq
|
Sorting
|
|
Astrocyte bulk RNA-seq
|
GSE73721
Ye Zhang et al. 2016
|
NextSeq500
|
-
|
Immunopanning
|
|
Astrocyte spheroid scRNA-seq
|
GSE99951
Sloan et al. 2017
|
NextSeq500
|
SMART-seq
|
Immunopanning
|
|
Developing human brain cortex
ScRNA-seq
|
dbGaP: phs000989.v3
Nowakowski et al. 2017
|
HiSeq2500
|
FluidigmC1
|
Stereotaxic microdissection
|
TableS3. Identities of cell clusters in developing human brain (derived from Nowakowski et al. 2017).
|
Cluster Name
|
Cluster Interpretation
|
|
Astrocyte
|
Astocyte
|
|
IN-CTX-CGE1
|
CGE/LGE-derived inhibitory neurons
|
|
IN-CTX-CGE2
|
CGE/LGE-derived inhibitory neurons
|
|
Choroid
|
Choroid
|
|
IPC-div1
|
Dividing Intermediate Progenitor Cells RG-like
|
|
MGE-div
|
dividing MGE Progenitors
|
|
RG-div1
|
Dividing Radial Glia (G2/M-phase)
|
|
RG-div2
|
Dividing Radial Glia (S-phase)
|
|
EN-PFC2
|
Early and Late Born Excitatory Neuron PFC
|
|
EN-PFC3
|
Early and Late Born Excitatory Neuron PFC
|
|
EN-V1-2
|
Early and Late Born Excitatory Neuron V1
|
|
EN-PFC1
|
Early Born Deep Layer/subplate Excitatory Neuron PFC
|
|
EN-V1-1
|
Early Born Deep Layer/subplate Excitatory Neuron V1
|
|
RG-early
|
earlyRG
|
|
Endothelial
|
Endothelial
|
|
EN-V1-3
|
Excitatory Neuron V1 - late born
|
|
Glyc
|
Glycolysis
|
|
IPC-nEN1
|
Intermediate Progenitor Cells EN-like
|
|
IPC-nEN2
|
Intermediate Progenitor Cells EN-like
|
|
IPC-nEN3
|
Intermediate Progenitor Cells EN-like
|
|
IPC-div2
|
Intermediate Progenitor Cells RG-like
|
|
nIN1
|
MGE newborn neurons
|
|
nIN2
|
MGE newborn neurons
|
|
nIN3
|
MGE newborn neurons
|
|
nIN4
|
MGE newborn neurons
|
|
nIN5
|
MGE newborn neurons
|
|
MGE-IPC1
|
MGE Progenitors
|
|
MGE-IPC2
|
MGE Progenitors
|
|
MGE-IPC3
|
MGE Progenitors
|
|
MGE-RG1
|
MGE Radial Glia 1
|
|
MGE-RG2
|
MGE Radial Glia 2
|
|
IN-CTX-MGE2
|
MGE-derived Ctx inhibitory neuron, Cortical Plate-enriched
|
|
IN-CTX-MGE1
|
MGE-derived Ctx inhibitory neuron, Germinal Zone Enriched
|
|
Microglia
|
Micrgolia
|
|
Mural
|
Mural/Pericyte
|
|
nEN-early2
|
Newborn Excitatory Neuron - early born
|
|
nEN-early1
|
Newborn Excitatory Neuron - early born
|
|
nEN-late
|
Newborn Excitatory Neuron - late born
|
|
OPC
|
Oligodendrocyte progenitor cell
|
|
oRG
|
Outer Radial Glia
|
|
IN-STR
|
Striatal neurons
|
|
tRG
|
Truncated Radial Glia
|
|
U1
|
Unknown1
|
|
U2
|
Unknown2
|
|
U3
|
Unknown3
|
|
U4
|
Unknown4
|
|
vRG
|
Ventricular Radial Glia
|
References
|
1. Fontenot, M. R. et al. Novel transcriptional networks regulated by CLOCK in human neurons. Genes Dev. 31, 2121-2135 (2017).
https://doi.org/10.1101/gad.305813.117 PMid:29196536 PMCid:PMC5749161
|
|
2. Goto, M. et al. Role of a circadian-relevant gene NR1D1 in brain development: possible involvement in the pathophysiology of autism spectrum disorders. Sci. Rep. 7, 43945 (2017). https://doi.org/10.1038/srep43945 PMid:28262759 PMCid:PMC5338261
|
|
3. Nicholas, B. et al. Association of Per1 and Npas2 with autistic disorder: support for the clock genes/social timing hypothesis. Mol. Psychiatry 12, 581-592 (2007). https://doi.org/10.1038/sj.mp.4001953 PMid:17264841
|
|
4. Li, P. et al. Loss of CLOCK Results in Dysfunction of Brain Circuits Underlying Focal Epilepsy. Neuron 96, 387-401.e6 (2017).
https://doi.org/10.1016/j.neuron.2017.09.044 PMid:29024662 PMCid:PMC6233318
|
| |
|
|
5. Gerstner, J. R. et al. BMAL1 controls the diurnal rhythm and set point for electrical seizure threshold in mice. Front. Syst. Neurosci. 8, 121 (2014). https://doi.org/10.3389/fnsys.2014.00121 PMid:25018707 PMCid:PMC4071977
|
|
6. Li, J. Z. et al. Circadian patterns of gene expression in the human brain and disruption in major depressive disorder. Proc. Natl. Acad. Sci. U. S. A. 110, 9950-9955 (2013). https://doi.org/10.1073/pnas.1305814110 PMid:23671070 PMCid:PMC3683716
|
| |
|
|
7. Bouchard-Cannon, P., Mendoza-Viveros, L., Yuen, A., Kærn, M. & Cheng, H.-Y. M. The circadian molecular clock regulates adult hippocampal neurogenesis by controlling the timing of cell-cycle entry and exit. Cell Rep. 5, 961-973 (2013). https://doi.org/10.1016/j.celrep.2013.10.037
PMid:24268780
|
|
8. Yang, G. et al. Timing of expression of the core clock gene Bmal1 influences its effects on aging and survival. Sci. Transl. Med. 8, 324ra16 (2016). https://doi.org/10.1126/scitranslmed.aad3305 PMid:26843191 PMCid:PMC4870001
|
|
9. Kobayashi, Y., Ye, Z. & Hensch, T. K. Clock genes control cortical critical period timing. Neuron 86, 264-275 (2015).
https://doi.org/10.1016/j.neuron.2015.02.036 PMid:25801703 PMCid:PMC4392344
|
|
10. Koike, N. et al. Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science 338, 349-354 (2012).
https://doi.org/10.1126/science.1226339 PMid:22936566 PMCid:PMC3694775
|
|
11. Ji, Z. & Ji, H. TSCAN: Pseudo-time reconstruction and evaluation in single-cell RNA-seq analysis. Nucleic Acids Res. 44, e117 (2016). https://doi.org/10.1093/nar/gkw430 PMid:27179027 PMCid:PMC4994863
|
|
12. Aibar, S. et al. SCENIC: single-cell regulatory network inference and clustering. Nat. Methods 14, 1083-1086 (2017).
https://doi.org/10.1038/nmeth.4463 PMid:28991892 PMCid:PMC5937676
|
|
13. Götz, M. & Huttner, W. B. The cell biology of neurogenesis. Nature Reviews Molecular Cell Biology vol. 6 777-788 (2005).
https://doi.org/10.1038/nrm1739 PMid:16314867
|
|
14. Petrik, D. & Encinas, J. M. Perspective: Of Mice and Men - How Widespread Is Adult Neurogenesis? Frontiers in Neuroscience vol. 13 (2019).
https://doi.org/10.3389/fnins.2019.00923 PMid:31555083 PMCid:PMC6727861
|
|
15. Yuzwa, S. A. et al. Developmental Emergence of Adult Neural Stem Cells as Revealed by Single-Cell Transcriptional Profiling. Cell Rep. 21, 3970-3986 (2017). https://doi.org/10.1016/j.celrep.2017.12.017 PMid:29281841
|
|
16. Hitzler, J. K. et al. Expression patterns of the hepatic leukemia factor gene in the nervous system of developing and adult mice. Brain Res. 820, 1-11 (1999). https://doi.org/10.1016/S0006-8993(98)00999-8
|
|
17. Rambousek, L. et al. Aberrant expression of PAR bZIP transcription factors is associated with epileptogenesis, focus on hepatic leukemia factor. Sci. Rep. 10, 3760 (2020). https://doi.org/10.1038/s41598-020-60638-7 PMid:32111960 PMCid:PMC7048777
|
|
18. Large-Scale Cellular-Resolution Gene Profiling in Human Neocortex Reveals Species-Specific Molecular Signatures. Cell 149, 483-496 (2012).
https://doi.org/10.1016/j.cell.2012.02.052 PMid:22500809 PMCid:PMC3328777
|
| |
|
19. Clark, E. A. et al. Cortical RORβ is required for layer 4 transcriptional identity and barrel integrity. Elife 9, (2020).
https://doi.org/10.7554/eLife.52370 PMid:32851975 PMCid:PMC7492084
|
|
20. Doetsch, F., García-Verdugo, J. M. & Alvarez-Buylla, A. Regeneration of a germinal layer in the adult mammalian brain. Proc. Natl. Acad. Sci. U. S. A. 96, 11619-11624 (1999). https://doi.org/10.1073/pnas.96.20.11619 PMid:10500226 PMCid:PMC18083
|
|
21. Kriegstein, A. & Alvarez-Buylla, A. The glial nature of embryonic and adult neural stem cells. Annu. Rev. Neurosci. 32, 149-184 (2009).
https://doi.org/10.1146/annurev.neuro.051508.135600 PMid:19555289 PMCid:PMC3086722
|
|
22. van Praag, H., Kempermann, G. & Gage, F. H. Running increases cell proliferation and neurogenesis in the adult mouse dentate gyrus. Nat. Neurosci. 2, 266-270 (1999). https://doi.org/10.1038/6368 PMid:10195220
|
|
23. Toda, T., Parylak, S. L., Linker, S. B. & Gage, F. H. The role of adult hippocampal neurogenesis in brain health and disease. Molecular Psychiatry vol. 24 67-87 (2019). https://doi.org/10.1038/s41380-018-0036-2 PMid:29679070 PMCid:PMC6195869
|
|
24. Llorens-Bobadilla, E. et al. Single-Cell Transcriptomics Reveals a Population of Dormant Neural Stem Cells that Become Activated upon Brain Injury. Cell Stem Cell 17, 329-340 (2015). https://doi.org/10.1016/j.stem.2015.07.002 PMid:26235341
|
|
25. Shin, J. et al. Single-Cell RNA-Seq with Waterfall Reveals Molecular Cascades underlying Adult Neurogenesis. Cell Stem Cell 17, 360-372 (2015). https://doi.org/10.1016/j.stem.2015.07.013 PMid:26299571
|
|
26. Liu, L. et al. Ndrg2 expression in neurogenic germinal zones of embryonic and postnatal mouse brain. J. Mol. Histol. 43, 27-35 (2012).
https://doi.org/10.1007/s10735-011-9378-7 PMid:22143493
|
|
27. Peña-Melian, A. Development of human brain. Hum. Evol. 15, 99-112 (2000). https://doi.org/10.1007/BF02436238
|
|
28. Silbereis, J. C., Pochareddy, S., Zhu, Y., Li, M. & Sestan, N. The Cellular and Molecular Landscapes of the Developing Human Central Nervous System. Neuron 89, 248-268 (2016). https://doi.org/10.1016/j.neuron.2015.12.008 PMid:26796689 PMCid:PMC4959909
|
|
29. Nowakowski, T. J. et al. Spatiotemporal gene expression trajectories reveal developmental hierarchies of the human cortex. Science 358, 1318-1323 (2017). https://doi.org/10.1126/science.aap8809 PMid:29217575 PMCid:PMC5991609
|
|
30. Haynes, S. E. et al. The P2Y12 receptor regulates microglial activation by extracellular nucleotides. Nat. Neurosci. 9, 1512-1519 (2006). https://doi.org/10.1038/nn1805 PMid:17115040
|
|
31. Lou, N. et al. Purinergic receptor P2RY12-dependent microglial closure of the injured blood-brain barrier. Proceedings of the National Academy of Sciences vol. 113 1074-1079 (2016). https://doi.org/10.1073/pnas.1520398113 PMid:26755608 PMCid:PMC4743790
|
|
32. Wirz, K. T. S. et al. Cortical beta amyloid protein triggers an immune response, but no synaptic changes in the APPswe/PS1dE9 Alzheimer's disease mouse model. Neurobiol. Aging 34, 1328-1342 (2013). https://doi.org/10.1016/j.neurobiolaging.2012.11.008 PMid:23245294
|
|
33. Chow, K.-H. et al. S100A4 Is a Biomarker and Regulator of Glioma Stem Cells That Is Critical for Mesenchymal Transition in Glioblastoma. Cancer Res. 77, 5360-5373 (2017). https://doi.org/10.1158/0008-5472.CAN-17-1294 PMid:28807938 PMCid:PMC5626628
|
|
34. Mohri, I. et al. Hematopoietic Prostaglandin D Synthase and DP1 Receptor Are Selectively Upregulated in Microglia and Astrocytes Within Senile Plaques From Human Patients and in a Mouse Model of Alzheimer Disease. Journal of Neuropathology and Experimental Neurology vol. 66 469-480 (2007). https://doi.org/10.1097/01.jnen.0000240472.43038.27 PMid:17549007
|
|
35. Li, H. et al. Transcription factor MEF2C influences neural stem/progenitor cell differentiation and maturation in vivo. Proc. Natl. Acad. Sci. U. S. A. 105, 9397-9402 (2008). https://doi.org/10.1073/pnas.0802876105 PMid:18599437 PMCid:PMC2453715
|
|
36. Zweier, M. et al. Mutations in MEF2C from the 5q14.3q15 microdeletion syndrome region are a frequent cause of severe mental retardation and diminish MECP2 and CDKL5 expression. Hum. Mutat. 31, 722-733 (2010). https://doi.org/10.1002/humu.21253 PMid:20513142
|
|
37. Shimozaki, K. Involvement of Nuclear Receptor REV-ERBβ in Formation of Neurites and Proliferation of Cultured Adult Neural Stem Cells. Cell. Mol. Neurobiol. 38, 1051-1065 (2018). https://doi.org/10.1007/s10571-018-0576-7 PMid:29397477
|
|
38. Mochizuki, T. et al. Functional investigation of solute carrier family 35, member F2 (SLC35F2) in three cellular models of the primate blood-brain barrier. Drug Metab. Dispos. (2020) doi:10.1124/dmd.120.000115. https://doi.org/10.1124/dmd.120.000115 PMid:33144341
|
|
39. Gao, P. P. et al. Regulation of topographic projection in the brain: Elf-1 in the hippocamposeptal system. Proc. Natl. Acad. Sci. U. S. A. 93, 11161-11166 (1996). https://doi.org/10.1073/pnas.93.20.11161 PMid:8855326 PMCid:PMC38301
|
|
40. Costa, L. G. et al. Paraoxonase-2 (PON2) in brain and its potential role in neuroprotection. Neurotoxicology 43, 3-9 (2014).
https://doi.org/10.1016/j.neuro.2013.08.011 PMid:24012887 PMCid:PMC3942372
|
|
41. Masuda, T. et al. IRF8 is a critical transcription factor for transforming microglia into a reactive phenotype. Cell Rep. 1, 334-340 (2012).
https://doi.org/10.1016/j.celrep.2012.02.014 PMid:22832225 PMCid:PMC4158926
|
|
42. Wehrspaun, C. C., Haerty, W. & Ponting, C. P. Microglia recapitulate a hematopoietic master regulator network in the aging human frontal cortex. Neurobiol. Aging 36, 2443.e9-2443.e20 (2015). https://doi.org/10.1016/j.neurobiolaging.2015.04.008 PMid:26002684 PMCid:PMC4503803
|
|
43. Galatro, T. F. et al. Transcriptomic analysis of purified human cortical microglia reveals age-associated changes. Nat. Neurosci. 20, 1162-1171 (2017). https://doi.org/10.1038/nn.4597 PMid:28671693
|
|
44. Tso, C. F. et al. Astrocytes Regulate Daily Rhythms in the Suprachiasmatic Nucleus and Behavior. Curr. Biol. 27, 1055-1061 (2017).
https://doi.org/10.1016/j.cub.2017.02.037 PMid:28343966 PMCid:PMC5380592
|
|
45. Lananna, B. V. et al. Cell-Autonomous Regulation of Astrocyte Activation by the Circadian Clock Protein BMAL1. Cell Rep. 25, 1-9.e5 (2018).
https://doi.org/10.1016/j.celrep.2018.09.015 PMid:30282019 PMCid:PMC6221830
|
|
|
46. Zhang, Y. et al. Purification and Characterization of Progenitor and Mature Human Astrocytes Reveals Transcriptional and Functional Differences with Mouse. Neuron vol. 89 37-53 (2016). https://doi.org/10.1016/j.neuron.2015.11.013 PMid:26687838 PMCid:PMC4707064
|
|
47. Sloan, S. A. et al. Human Astrocyte Maturation Captured in 3D Cerebral Cortical Spheroids Derived from Pluripotent Stem Cells. Neuron 95, 779-790.e6 (2017). https://doi.org/10.1016/j.neuron.2017.07.035 PMid:28817799 PMCid:PMC5890820
|
|
48. He, Y. et al. The transcriptional repressor DEC2 regulates sleep length in mammals. Science 325, 866-870 (2009).
https://doi.org/10.1126/science.1174443 PMid:19679812 PMCid:PMC2884988
|
|
49. Pellegrino, R. et al. A novel BHLHE41 variant is associated with short sleep and resistance to sleep deprivation in humans. Sleep 37, 1327-1336 (2014). https://doi.org/10.5665/sleep.3924 PMid:25083013 PMCid:PMC4096202
|
|
50. Zhang, Y. et al. An RNA-Sequencing Transcriptome and Splicing Database of Glia, Neurons, and Vascular Cells of the Cerebral Cortex. Journal of Neuroscience vol. 34 11929-11947 (2014). https://doi.org/10.1523/JNEUROSCI.1860-14.2014 PMid:25186741 PMCid:PMC4152602
|
|
51. Li et al. GFAP Mutations in Astrocytes Impair Oligodendrocyte Progenitor Proliferation and Myelination in an hiPSC Model of Alexander Disease. Cell Stem Cell vol. 23 239-251.e6 (2018). https://doi.org/10.1016/j.stem.2018.07.009 PMid:30075130 PMCid:PMC6230521
|
|
52. Liu, Y. et al. CD44 expression identifies astrocyte-restricted precursor cells. Dev. Biol. 276, 31-46 (2004).
https://doi.org/10.1016/j.ydbio.2004.08.018 PMid:15531362
|
|
53. Akiyama, H., Tooyama, I., Kawamata, T., Ikeda, K. & McGeer, P. L. Morphological diversities of CD44 positive astrocytes in the cerebral cortex of normal subjects and patients with Alzheimer's disease. Brain Res. 632, 249-259 (1993). https://doi.org/10.1016/0006-8993(93)91160-T
|
|
54. Seney, M. L. et al. Diurnal rhythms in gene expression in the prefrontal cortex in schizophrenia. Nat. Commun. 10, 3355 (2019).
https://doi.org/10.1038/s41467-019-11335-1 PMid:31399567 PMCid:PMC6689017
|
|