Abstract microRNAs (miRNAs) are small non-coding RNAs ~22nt in length that regulate gene expression post transcriptionally by binding to their 3’UTR. In the last 27 years, since its discovery, tremendous progress has been made to determine its functional significance in the spatio-temporal regulation of gene expression; they act cordially to regulate gene networks or pathways. miRNA being stable, are also found in body fluid serving as a potential biomarker. Despite knowing a lot about miRNAs function, its use in clinical phase is still in its infancy- with poor outcome in clinical trials and difficulty in its tissue-specific delivery. This review provides a snapshot of the progress of miRNA field in past few years and challenges associated with its clinical applications.
Keywords: miRNA, targeted delivery, clinical trials, gene expression
Background
The discovery of microRNAs (miRNAs) marked the beginning of an era of designating role to otherwise considered junk non-coding RNAs. These small non-coding RNAs, called microRNAs, control the gene expression during development and disease; and are one of the major mechanisms of spatio-temporal regulation. This further leads to the discovery of other non-coding RNAs, such as long non-coding RNAs, circular RNAs, and piwi RNAs(1,2). This review solitary focuses on miRNAs, how its field has progressed through 27 years and what are the challenges associated with their therapeutic usage.
In 1993, two articles were published back-to-back by Ambros and Ruvkun team in Cell(3,4), demonstrating the post-transcriptional regulation of lin-14 gene in C. elegans development by a non-coding RNA- lin-4 for the first time. From their earlier studies, they found that LIN-14 protein shows temporal expression at different stages of C. elegans development without change in its mRNA, indicating post-transcriptional regulation. They found that lin-4 negatively regulates LIN-14 and the regulatory elements were present in the lin-14 3’UTR as determined by deletion experiments. However, the mechanistic details were not known. Both groups independently discovered in 1993 that lin-4 exists as 22nt and 61nt long fragment, which we now know as mature and precursor miRNA that acts by binding to the complementary regions in 3’UTR of lin-14.
Surprisingly, there was no other miRNA study published for seven years. Then both teams came together and published another miRNA, namely let-7 that was found to inhibit the inhibitor - lin-41, which in turn blocks the action of LIN-29 - an adult specification transcription factor in C. elegans, thus relieving the lin-41 inhibition5 and controlling adult development. Further, Ruvkun and team found that let-7 was conserved among various species including vertebrates demonstrating its expression in various human tissues6. The discovery of miRNA in humans led to a spike in the studies on miRNAs in human development and disease, along with development of bioinformatic tools to build miRNA database, predicting genes encoding miRNAs and their targets. Interestingly, the number of miRNAs in a complex organism, such as humans, were found to be higher when compared to plants or flies, indicating their evolutionary role and mechanism of regulating complicated organisms(7-9). Thus, the secret lies in the non-coding part of the genome that controls the protein coding genes enabling them for diverse functions.
The first miRNA study in human disease was in 2002, where Calin et al. found that miR-15 and miR-16 regions were deleted in B-cell chronic lymphocytic leukemia from peripheral blood samples of patients(10). Thus, it took about 10 years for the miRNA field to progress from C. elegans to humans. However, they did not focus on molecular mechanism to speculate any causative link between the miRNA and leukemia in this study. Many other studies also determined the role of miRNAs in human disease and development, which is thoroughly reviewed by Ardekani and Naeini(11). Further, Sayed and Abdellatif comprehensively reviewed miRNAs involved in various biological processes such as hematopoiesis, cardiac hypertrophy, cardiac ischemia, myogenesis, glioblastoma and various kind of cancers(12). This article provides a beautiful representation of these pathways. Looking closer into the effects of these miRNAs, it was observed that some miRNA, such as miR-21, are pleiotropic in their action and regulate multiple processes, such as hematopoiesis, cardiac fibrosis and cancer. Such miRNAs are difficult to target due to their diverse actions in different tissues. On the other hand, tissue-specific miRNA could be an attractive therapeutic target, such as miRNA cluster consisting of miR-338–3p, miR-219a-5p, miR-124–3p and miR-9–5p that is brain specific and miR-507, miR-514a-3p and miR-509–5p that are expressed exclusively in testis. Beyond these, there are single miRNAs, such as miR-122-5p that are expressed only in the lungs(13). As determined from miRNA atlas, about 83.7% of miRNAs are abundant across all tissues; thus, care should be taken when choosing miRNAs for therapeutic application, encouraging tissue-specific labelling. The detailed miRNA profiling of each human tissue is available on miRNA atlas- http://ccb-web.cs.uni-saarland.de/tissueatlas. Overall timeline of miRNA discovery is represented in Figure 1.
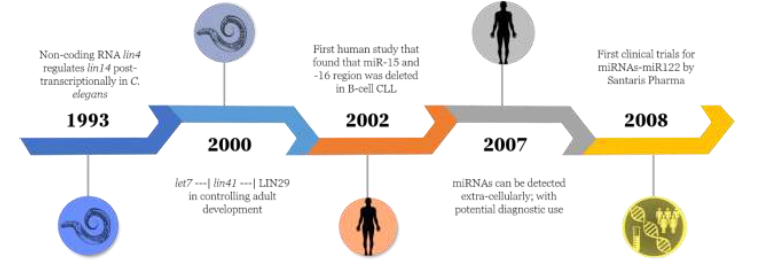
Figure 1. Timeline of miRNA discovery from C. elegans to clinical trials in humans.
Furthermore, many molecular mechanisms exist to control miRNA expression and function(14). Some of them generate isomiRs- that are miRNAs differing in sequence or length due to nucleotide addition, deletion at 5’ or 3’ termini, or substitution. The nucleotides can be deleted by template dependent exonuclease and can be added by non-template dependent nucleotidyl transferase. The substitution can occur by A-to-I or C-to-U editing of miRNA by ADAR (Adenosine deaminase acting on RNA) and APOBEC (apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like) enzymes respectively(15,16). These isomiRs can in turn affect the stability and binding ability of miRNA to its targets. For example, editing occurring in the seed region can alter its target binding ability, thereby, binding to a completely new set of targets. Increased miRNA editing has been associated with cancer(17) and brain development(18,19). All these studies indicate potential use of edited miRNA as a biomarker and therapeutic tool.
Other ways of affecting miRNA expression include regulation of proteins involved in miRNA biogenesis, such as Ago, Drosha, Dicer, exportin etc. Intronic miRNA-expressing within a host gene, can follow its host gene expression. Thus, factors regulating the host genes can in turn affect miRNA and alter its downstream mediators. miRNA expression can also be regulated epigenetically by altering its DNA or histone methylation and acetylation. Lastly, many exogenous factors can modify miRNA expression patterns, such as stress, steroid hormones, hypoxia, carcinogens, and so on(20, 21). One example of mediating effects of hypoxia is by HIF-1α, which is a transcription factor and controls various miRNAs directly and indirectly(22). Similarly, many studies have predicted the relationship between transcription factor and miRNAs, hence, forming an intricate network(23, 24).
In addition to therapeutic targets, miRNAs serve as a biomarker as they are stable, and are easily detected in tissue fluid as free form or enclosed in vesicles. In 2007, Valadi et al. were the first to demonstrate that miRNAs can communicate between cells via exosomes in mast cells(25). Later, Chen et al. in 2008 also showed presence of miRNA in serum where they found increased levels of miR-25 and miR-223 in lung cancer (n=152) as compared to healthy control (n=75)(26). Furthermore, numerous studies demonstrated altered levels of miRNAs in patients’ sera of various diseases and also as a mode of communication between cells that is reviewed extensively in the literature(27-29). The major challenges associated with miRNAs as biomarker is the absence of proper normalization control, usage of small sample size, not characterizing patients’ samples in terms of sub-classification of the disease and including other clinical co-morbidities(30). Thus, the use of a universal method for miRNA isolation and normalization would be a better strategy for identification of miRNAs as biomarkers to apply to a bigger population. For example, Faraldi et al. has found hsa-miR-320d as a reference miRNA by using various normalization strategies to overcome this problem(31); thus, many studies like this are encouraged. Having a miRNA-based committee might be helpful in establishing a gold standard method for miRNA research to get a fruitful outcome.
Studies focused on miRNA biology follow a very standardized approach as shown in Figure 2. The first step is to discover a miRNA(s)-gene target pair(s) using bioinformatics prediction softwares that are mostly based on sequence complementarity between the miRNA seed region and 3’UTR site. Chen et al. have compiled a list of bioinformatics tools that assist in identification of miRNAs, their sequence and pre-miR structure, target prediction- both computationally and experimentally, and miRNA interaction network to other miRNA, and so on32. The main focus has been given to target prediction tools that are based on different principles, such as sequence complementarity to the seed region (TargetScan), self-organization map (mirSOM), or experimentally validated miRNA (DIANE TarBase). Once a miRNA and its target gene(s) are identified, their binding is validated by 3’UTR luciferase promoter assay, followed by gain and loss of experiments in both cellular and animal models to determine their biological function; followed by testing their translational ability in human samples. According to miRBase, 1917 human miRNAs have been identified till now, but only a few have made their way to clinical validation and trials. This is possibly due to diverse effect of miRNAs on many genes/pathways, making it difficult to use therapeutically. Thus, to make progress in miRNA field, care should be given towards its validation in human studies and improving its tissue-specific delivery. Many modes of delivery of miRNAs are available that carry modified miRNAs mimics or antagomirs to prevent their degradation. They are further enclosed into vesicles made from polymer, lipid, viral vector, membrane vesicles, nanoparticles, etc. for in vivo delivery and uptake by tissue(33,34). They are sometimes labeled by ligands to take up by specific cell receptors, but such studies are scarce. Thus, more focus should be given to cell specific delivery of miRNAs to overcome off-target effects.
Figure 2. Standardized approach for miRNA study.
A miRNA study is begun by first identifying the miRNA or gene of interest and finding its target using bioinformatics approach. Followed by in vitro binding using luciferase assay and gain and loss of function in cellular and animal model of disease. Lastly validating its human relevance by determining its expression or function in patients’ samples.
Figure 3. Distribution of miRNAs clinical trials as of Oct 2020.
The clinical trials website listed 945 trials when searched for ‘miRNA’ as input. This pie chart shows the distribution of them.
Lastly, we searched the keyword ‘miRNA’ in the clinical trials database (https://clinicaltrials.gov/) to determine the progress of miRNA and found 945 trials, as of Oct 2020. The distribution of these trials in terms of their status is shown in Figure 3. Among these, 488 were observational and 457 interventional; interestingly in interventional studies, most of them were based on miRNAs as diagnostic marker, very few included drugs that altered miRNA expression and function. The status of most of the interventional studies was unknown at that time, only one of them reported their outcome as an adverse effect. This study was the phase-I trial of administration of miR34a enclosed in liposome vesicle in patients with advanced solid tumors. The trial was closed early due to immune related adverse outcomes and death of 4 patients(35), indicating infancy of miRNA in clinics. Thus, cell specific delivery and uptake of miRNAs and protection from off-target immune responses should be the focus of future studies. Another strategy to improve the translational potential is the use of an appropriate animal model that mimics actual disease conditions or utilization of different models based on different molecular mechanisms of disease. This could assist in improved outcome of miRNAs therapy. The dynamic regulation of miRNAs is difficult to comprehend, their differential regulation in tissues makes miRNAs therapeutic a blessing in disguise. Thus, the progress from bench to bedside could be possible if the process of miRNAs discovery and development could be streamlined.
REFERENCES
01. Beermann, J., et al., Non-coding RNAs in Development and Disease: Background, Mechanisms, and Therapeutic Approaches. Physiol Rev, 2016. 96(4): p. 1297-325. https://doi.org/10.1152/physrev.00041.2015 PMid:27535639
02.Ozata, D.M., et al., PIWI-interacting RNAs: small RNAs with big functions. Nat Rev Genet, 2019. 20(2): p. 89-108.
https://doi.org/10.1038/s41576-018-0073-3 PMid:30446728
03. Lee, R.C., R.L. Feinbaum, and V. Ambros, The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell, 1993. 75(5): p. 843-54. https://doi.org/10.1016/0092-8674(93)90529-Y
04. Wightman, B., I. Ha, and G. Ruvkun, Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell, 1993. 75(5): p. 855-62. https://doi.org/10.1016/0092-8674(93)90530-4
05. Slack, F.J., et al., The lin-41 RBCC gene acts in the C. elegans heterochronic pathway between the let-7 regulatory RNA and the LIN-29 transcription factor. Mol Cell, 2000. 5(4): p. 659-69. https://doi.org/10.1016/S1097-2765(00)80245-2
06. Pasquinelli, A.E., et al., Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature, 2000. 408(6808): p. 86-9. https://doi.org/10.1038/35040556 PMid:11081512
07. Heimberg, A.M., et al., MicroRNAs and the advent of vertebrate morphological complexity. Proc Natl Acad Sci U S A, 2008. 105(8): p. 2946-50.
https://doi.org/10.1073/pnas.0712259105 PMid:18287013 PMCid:PMC2268565
08. Dexheimer, P.J. and L. Cochella, MicroRNAs: From Mechanism to Organism. Front Cell Dev Biol, 2020. 8: p. 409.
https://doi.org/10.3389/fcell.2020.00409 PMid:32582699 PMCid:PMC7283388
09. Moran, Y., et al., The evolutionary origin of plant and animal microRNAs. Nat Ecol Evol, 2017. 1(3): p. 27. https://doi.org/10.1038/s41559-016-0027 PMid:28529980 PMCid:PMC5435108
10. Calin, G.A., et al., Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A, 2002. 99(24): p. 15524-9. https://doi.org/10.1073/pnas.242606799 PMid:12434020 PMCid:PMC137750
11. Ardekani, A.M. and M.M. Naeini, The Role of MicroRNAs in Human Diseases. Avicenna J Med Biotechnol, 2010. 2(4): p. 161-79. PMid:23407304 PMCid:PMC3558168
12. Sayed, D. and M. Abdellatif, MicroRNAs in development and disease. Physiol Rev, 2011. 91(3): p. 827-87. https://doi.org/10.1152/physrev.00006.2010 PMid:21742789
13. Ludwig, N., et al., Distribution of miRNA expression across human tissues. Nucleic Acids Res, 2016. 44(8): p. 3865-77. https://doi.org/10.1093/nar/gkw116 PMid:26921406 PMCid:PMC4856985
14. Gebert, L.F.R. and I.J. MacRae, Regulation of microRNA function in animals. Nat Rev Mol Cell Biol, 2019. 20(1): p. 21-37.
https://doi.org/10.1038/s41580-018-0045-7 PMid:30108335 PMCid:PMC6546304
15. Jasdeep Kaur Dhanoa, R.V., R. S. Sethi, Jaspreet Singh Arora & C. S. Mukhopadhyay, Biogenesis and biological implications of isomiRs in mammals- a review. ExRNA, 2019. 1. https://doi.org/10.1186/s41544-018-0003-8
16. Neilsen, C.T., G.J. Goodall, and C.P. Bracken, IsomiRs--the overlooked repertoire in the dynamic microRNAome. Trends Genet, 2012. 28(11): p. 544-9. https://doi.org/10.1016/j.tig.2012.07.005 PMid:22883467
17. Pinto, Y., et al., Human cancer tissues exhibit reduced A-to-I editing of miRNAs coupled with elevated editing of their targets. Nucleic Acids Res, 2018. 46(1): p. 71-82. https://doi.org/10.1093/nar/gkx1176 PMid:29165639 PMCid:PMC5758889
18. Ekdahl, Y., et al., A-to-I editing of microRNAs in the mammalian brain increases during development. Genome Res, 2012. 22(8): p. 1477-87.
https://doi.org/10.1101/gr.131912.111 PMid:22645261 PMCid:PMC3409261
19. Mingardi, J., et al., miRNA Editing: New Insights into the Fast Control of Gene Expression in Health and Disease. Mol Neurobiol, 2018. 55(10): p. 7717-7727. https://doi.org/10.1007/s12035-018-0951-x PMid:29460265
20. Gulyaeva, L.F. and N.E. Kushlinskiy, Regulatory mechanisms of microRNA expression. J Transl Med, 2016. 14(1): p. 143.
https://doi.org/10.1186/s12967-016-0893-x PMid:27197967 PMCid:PMC4873990
21. Schanen, B.C. and X. Li, Transcriptional regulation of mammalian miRNA genes. Genomics, 2011. 97(1): p. 1-6.
https://doi.org/10.1016/j.ygeno.2010.10.005 PMid:20977933 PMCid:PMC3019299
22. Kulshreshtha, R., et al., A microRNA signature of hypoxia. Molecular and Cellular Biology, 2007. 27(5): p. 1859-1867.
https://doi.org/10.1128/MCB.01395-06 PMid:17194750 PMCid:PMC1820461
23. Martinez, N.J. and A.J. Walhout, The interplay between transcription factors and microRNAs in genome-scale regulatory networks. Bioessays, 2009. 31(4): p. 435-45. https://doi.org/10.1002/bies.200800212 PMid:19274664 PMCid:PMC3118512
24. Wang, H., et al., Investigating MicroRNA and transcription factor co-regulatory networks in colorectal cancer. BMC Bioinformatics, 2017. 18(1): p. 388. https://doi.org/10.1186/s12859-017-1796-4 PMid:28865443 PMCid:PMC5581471
25. Valadi, H., et al., Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol, 2007. 9(6): p. 654-9. https://doi.org/10.1038/ncb1596 PMid:17486113
26. Chen, X., et al., Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res, 2008. 18(10): p. 997-1006. https://doi.org/10.1038/cr.2008.282 PMid:18766170
27. He, B., et al., miRNA-based biomarkers, therapies, and resistance in Cancer. Int J Biol Sci, 2020. 16(14): p. 2628-2647.
https://doi.org/10.7150/ijbs.47203 PMid:32792861 PMCid:PMC7415433
28. Etheridge, A., et al., Extracellular microRNA: a new source of biomarkers. Mutat Res, 2011. 717(1-2): p. 85-90.
https://doi.org/10.1016/j.mrfmmm.2011.03.004 PMCid:PMC3199035
29. Mori, M.A., et al., Extracellular miRNAs: From Biomarkers to Mediators of Physiology and Disease. Cell Metab, 2019. 30(4): p. 656-673. https://doi.org/10.1016/j.cmet.2019.07.011 PMid:31447320 PMCid:PMC6774861
30. Condrat, C.E., et al., miRNAs as Biomarkers in Disease: Latest Findings Regarding Their Role in Diagnosis and Prognosis. Cells, 2020. 9(2). https://doi.org/10.3390/cells9020276 PMid:31979244 PMCid:PMC7072450
31. Faraldi, M., et al., Normalization strategies differently affect circulating miRNA profile associated with the training status. Sci Rep, 2019. 9(1): p. 1584. https://doi.org/10.1038/s41598-019-38505-x PMid:30733582 PMCid:PMC6367481
32. Chen, L., et al., Trends in the development of miRNA bioinformatics tools. Brief Bioinform, 2019. 20(5): p. 1836-1852.
https://doi.org/10.1093/bib/bby054 PMid:29982332 PMCid:PMC7414524
33. Yong Fu, J.C., Zhen Huang, Recent progress in microRNA-based delivery systems for the treatment of human disease. ExRNA, 2019. 1.
https://doi.org/10.1186/s41544-019-0024-y
34. Bajan, S. and G. Hutvagner, RNA-Based Therapeutics: From Antisense Oligonucleotides to miRNAs. Cells, 2020. 9(1).
https://doi.org/10.3390/cells9010137 PMid:31936122 PMCid:PMC7016530
35. Hong, D.S., et al., Phase 1 study of MRX34, a liposomal miR-34a mimic, in patients with advanced solid tumours. Br J Cancer, 2020. 122(11): p. 1630-1637. https://doi.org/10.1038/s41416-020-0802-1 PMid:32238921
|