Abstract
Caenorhabditis elegans infection by Orsay virus initiates a variety of abnormalities, including intestinal damages. However, the role of the viral capsid proteins to this pathogenicity needs to be further discovered. To address this question, engineered bacterial lawn expressing recombinant viral proteins were used as a food source to deliver them to the worm gut, and survival and behavioral studies by assessing worm mobility were performed. Worm fed with protein α-δ and δ survived longer while δ contributes to worm intestinal defects. Morphological analysis suggests that protein α limits food absorption in the intestine while α-δ plays a role in food digestion. Moreover, experiments tracking worm mobility showed hyperactive behavior by more forward and less reverse movement, as well as less time pausing. It shows that capsid proteins promote an adaptive behavioral response to food ingestion and locomotion. Since hyperactivity has been previously linked to serotonin by others, the viral protein may modulate the serotonin level in the worm gut. These data are consistent with comparative modeling results of capsid proteins showing that protein delta has a high degree of structural similarity with the rate-limiting enzyme (TDO-2) of the kynurenine pathway. Because TDO-2 controls serotonin production and promotes brain excitability, we can hypothesize that protein δ mimics and/or interferes with host TDO-2 activity. Since hyperactivity has been reported in other virus-human systems, it could be that this behavioral response is evolutionary conserved – a possibility that require further exploration. Keywords: Virus-Host interaction, Orsay capsid proteins, Pathogenesis, Life span, Food behavior, Locomotion
1. Introduction
Discovered almost a decade ago, Orsay Virus (OrV) is the only known virus capable of naturally infecting the model organism Caenorhabditis elegans (C. elegans) 1. OrV is a bipartite positive-sense single stranded RNA virus related to family Nodaviridae that expresses a total of four polypeptides, including an RNA-dependent RNA polymerase (RdRP), a capsid protein-alpha (CP-α), a non-structural protein delta (δ), and a fusion protein capsid-delta (CP-δ/α-δ) produced by ribosomal frameshift 2. Orsay primarily infects the nematodes digestive tract causing abnormal intestinal symptoms without killing the worms 1. RdRP functions in viral transcription and replication, while the CP-α is subsequently cleaved into beta and gamma peptides during virus maturation 2. Similarly, the capsid-delta (CP-δ) protruding from the virus envelope has been shown to function likely by binding to the cell surface, allowing infection, but other functions have been suggested for CP-δ(3, 4). Consequently, further functional characterization of CPs during Orsay infection in the worm remains to be explored. Viral Capsid Proteins (CPs), the most abundant proteins made during viral infection, are not only mediating the entry of viral genome in or out of cells, but may play a significant role in virus infectivity and pathogenicity, contributing consequently to disease phenotypes(5, 6). Significant evidence in plant systems has contributed to our understanding of the regulatory function of CPs in host responses to viral infectivity in animals, in general. A better understanding of CPs contribution to host gene expression and antiviral immunity could reveal how metabolic pathways are rewired at the molecular level in the infected hosts 7. Since a virus-host system has been established in C. elegans infected by Orsay virus (OrV) and given the ease of performing experiments due to worm transparency to study intestinal function, we can now address how these viral proteins interfere with and reprogram the host machinery. In this study, we developed a not yet available experimental system that would enable to study the delivery of individual viral protein to the worm gut and further assess their contribution to host innate antiviral responses using subsequent downstream assays.
To survive and multiply inside a host, viruses perform complex procedures to reprogram host signaling, regulatory and metabolic machinery in a sustainable way. Among invertebrates, two basic mechanisms are being used by the host: The first is RNA interference (RNAi) silencing, an evolutionarily conserved system that neutralizes viral nucleic acids inside the cell and activates innate immunity pathways 8. The second mechanism uses protein produced by virus, that may be very similar in structure and function to some host proteins, allowing them to interact or interferes with host proteins and consequently reprogramming the host cellular machinery 9. The RNAi pathway plays an essential role in the C. elegans antiviral response. The inactivation of genes in the RNAi pathway such as rde-1 1 and drh-1 (10,11) can sensitize the wildtype N2 strain (which supports mild infection) to Orsay. In C. elegans, RNAi response to infection is neither systemic, nor there is evidence of transgenerational inheritance of silencing by Orsay (12), which is different from the RNAi induced by exogenous dsRNA. Additionally, the ubiquitin-mediated defence also supports Orsay resistance in worms (13).
C. elegans eats bacteria and feeds almost continuously. To detect food, they move toward it and use pharyngeal pumping that grinds then ingests bacteria. Worms can make elaborate decisions in the context of feeding. Two types of locomotory behavior occur in response to food, “dwelling” where the worm likes the food and stays, or “roaming” where the worm tries to find a more edible food source by moving away (14). These behavioral states are controlled by signaling from serotonin, insulin and transforming growth factor-β (TGF-β) (15). Dwelling corresponds to a reduced average translational speed but results in an increase in turning frequency, limiting the worm to a restricted area of food. Roaming has a higher average speed but fewer turning events, allowing exploration of a larger food patch. These behaviors are due to an exploration-exploitation decision that assesses food quality. As a result, the worm spends more time roaming and less time dwelling when the quality of the food is low (16). Conversely, when worms are on good quality food, roaming is rare (17). These food-related decisions are enabled by the nervous system that makes connections between sensory and mechano-sensory neurons and the gut (18,19). Those connections responsible for pharynx pumping and behavioral locomotion through the alimentary system are mainly controlled by 5-hydroxytryptamine (5-HT). The modulation of the behavioral response to food by 5-HT, results in an enhancement in pumping rate and reduction in locomotion (20). In C. elegans, nutritional availability is directly linked to food behavior and can be sensed by the rate-limiting enzyme Tryptophan 2,3-dioxygenase (TDO in human, TDO-2 in C. elegans) in the serotonin-kynurenine pathway that affects serotonin and kynurenic acid levels that results in different excitability states and lifespan extension in response to food cues (21, 22).
Classical reverse-genetics tools have been developed in the worm to study different viruses such as the Flock house virus, Pariacoto virus, and a fish-infecting Betanodavirus(23) and recently reported for the OrV 4). This does not provide a direct readout of the role of individual viral proteins since the RNAi response masks their contribution to the diseased phenotype. However, since C. elegans feeds on Escherichia coli (E. coli) OP50 and use them as a primary food source; it is relatively straightforward to perform studies of intestinal infections in this host model(24). This creates the opportunity to generate E. coli lawn expressing recombinant proteins of interest and to use them as a food source to deliver those specific proteins to the gut of the worm. This approach considers the bacterial lawn of C. elegans not only as a food source, but also as a genetically engineered microbiota environment to study host-pathogen interaction in the gut of the nematode.
To better define the functional role of Orsay capsid α, δ and α-δ during infection, a new method so-called PROtein FEeding in C. Elegans (PROFECE) was developed. Figure 1 shows a diagram listing the steps of this method. Worms are fed directly on a bacterial lawn expressing the recombinant protein of interest and as a result, delivers those proteins directly to the gut. This study was initially designed to investigate the contribution of capsid proteins to virus pathogenicity. However, the results also provided novel findings of the role of these proteins in extending the worm lifespan and an eye-opening clue to modulating worm locomotory behavior.
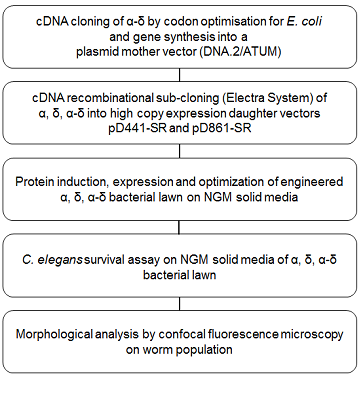
Figure 1. PROtein FEeding in C. Elegans (PROFECE) method.
2. Results
2.1. Optimized cDNA α-δ fusion produces high yield of folded and soluble protein
The optimized cDNA sequence of α-δ fusion was integrated into the bacterial protein expression vector pD441-SR, which initially allowed a strong lac promotor expression under the control of IPTG to ensure a high level of protein expression. As expected, the preliminary results (Figure 2) obtained on a small-scale protein expression study revealed that a large amount of protein was produced (340 µg ml-1) but with a significant fraction of this protein being insoluble or present as inclusion bodies. This indicated that the protein was produced too fast making inclusion bodies due to too much-misfolded proteins accumulating within the cell.
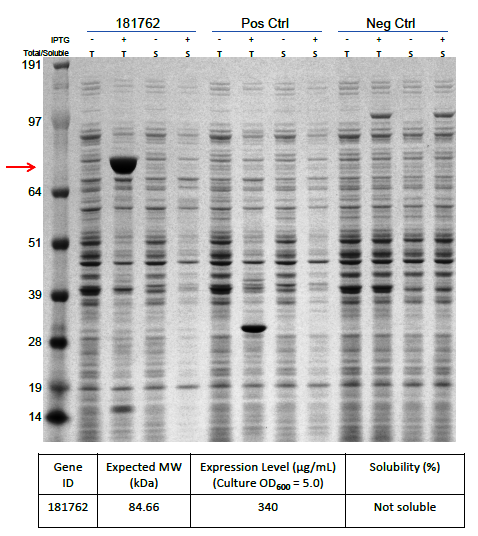
Figure 2. Protein expression of α-δ fusion protein after codon optimization.
Protein α-δ fusion is over-expressed in E. coli (340 mg/mL). T: Total fraction, S: Soluble fraction. 181762: Orsay virus capsid α-δ fusion protein. Neg. Ctrl: No -vector containing strain. Pos. Ctrl: Vector only strain. Red arrow indicates the expected molecular weight of α-δ fusion. Gel: 4-12% NuPAGE Bis-Tris (Invitrogen), buffered with MOPS containing reducing agent (Invitrogen), and stained with the SilverXpress silver staining Kit (Invitrogen). Ladder: Protein standard marker ranging from 10 to 260 kDa (Invitrogen).
We subsequently used BL21 Arctic cells to slow down the process. BL21 Arctic cells have shown an ability to optimize protein expression even at a temperature where E. coli is not functioning (4 °C). Figure 3 shows results from expression solubility experiments by SDS-PAGE of capsid protein α-δ on BL21 Arctic cells. Figure 3a shows the effect of (No RFP) not fused, or (RFP) fused to FresnoRFP at 37 ºC upon different conditions, (-) before induction and (+) after induction with IPTG. Figure 3b compares the results as a function of temperature from 11.5 ºC to 37 ºC. Obviously, the protein was more soluble at 20 °C than at 37 °C, the worm growth temperature.
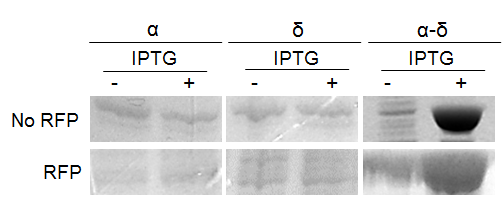
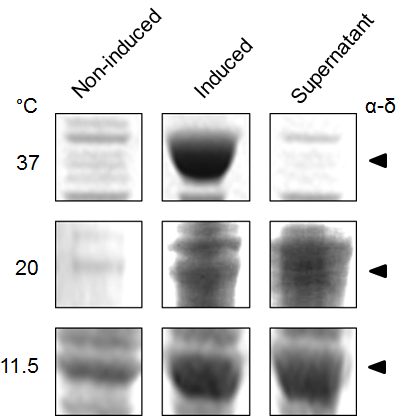
Figure 3. Expression of Orsay virus capsid protein α-δ.
(a) SDS-PAGE analysis of Orsay capsid proteins, No RFP: not fused FresnoRFP, RFP: fused to FresnoRFP at 37 ºC. Total lysate expression of α, δ, and α-δ protein in arctic cells. Protein normalization by optical density (280 nm). The expression was performed in different conditions, (-) before induction and (+) after induction with IPTG. (b) The capsid protein α-δ is more soluble at lower growth temperature. Protein expression was performed for 18 hours induction with 1mM IPTG.
The secretion and expression of the protein in NGM media was detected via the pink color, which could be seen visually in the media (Supplemental Figure S1), using the protein fused to FresnoRFP. The fused proteins were engineered by adding the RFP tag at the carboxy-terminal end (C terminus). A pink coloration of the bacterial growth in NGM media confirmed that the whole fusion protein, including the capsid region was folded properly (25). This opened the possibility to use the engineered BL21 Arctic cells as a food source for C. elegans. The solubility test results showed a higher solubility at lower temperature since the band in SDS PAGE was stronger in the supernatant fraction. Thereby, more proteins were expressed at a higher temperature; particularly true at 37 °C where the bands are thickest (Figure 3b). An important part of the protein stayed in the pellet, underlying the fact that the protein was insoluble, and probably formed inclusion bodies.
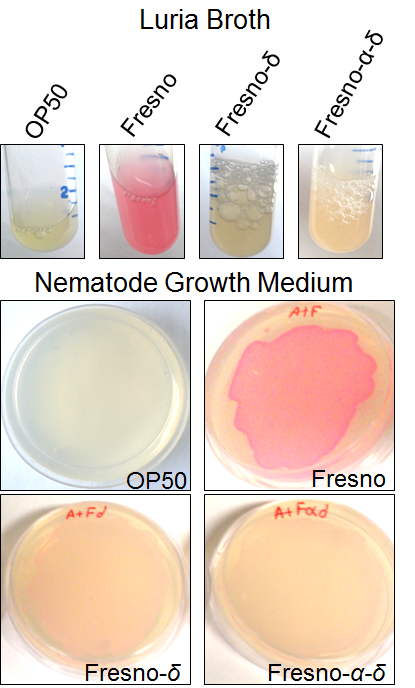
Supplemental Figure S1. Expression of RFP α-δ and δ proteins. Lawn of E. coli expressing capsid proteins fused to RFP can be grown on NGM (Nematode Growth Media) plates.
2.2. Protein δ extends lifespan of C. elegans
Figure 4 shows survival curve assays for C. elegans fed with different types of food. Survival curves of C. elegans fed with BL21 Arctic cells or the standard OP50 are compared in Figure 4a. A noteworthy difference was seen for the median survival time between OP50 than for BL21 arctic cells, however, the log rank analysis indicated that the difference is not statistically substantial (P = 0.51), suggesting that BL21 can be used as a better food source. This could be explained by the sample size in our experimental condition. However, since OP50 does not provides experimental system for the protein expression, BL21 arctic cells can be used still since it has a wide range of possible protein expression temperature as well as an engineered genetic system that allows the production of more stable recombinant protein at high yield. Figure 4b shows a comparison of results analyzed from C. elegans fed with BL21 arctic cells expressing proteins α-δ, α, and δ as compared to worms fed with BL21 Arctic cells only (n = 30, in triplicate). The median survival time was 1.5 days longer for worms fed with α-δ (P = 8x10-4). The lifespan of worms fed with protein δ was also lengthened by 1.5 days (P = 1.8x10-3). However, the nematodes fed with protein α showed a normal lifespan (P = 0.57) suggesting that δ contributed the most to worm’s lifespan extension. The extended lifespan data indicated that the protein α-δ food source is non-toxic. These results indicate that the worm lifespan can be regulated by dietary adaptation and caloric restriction by activating an immune response, consistent with previous studies (26, 27).
In addition, survival assay of protein α-δ lawn was assessed in the C. briggsae model, which is a non-specific host for OrV. Results plotted in Figure 4c show that protein α-δ affects worm lifespan in the same way as for C. elegans. This result indicates that α-δ proteins affect lifespan by a similar mechanism in both model organisms that is protein specific rather than host specific. It is consistent with the observations that capsid d protein are the most conserved proteins among the three known noda-like viruses in Caenorhabditis nematodes (Santeuil, Leblanc, Orsay) and share the same tissue tropism towards host intestinal cells.
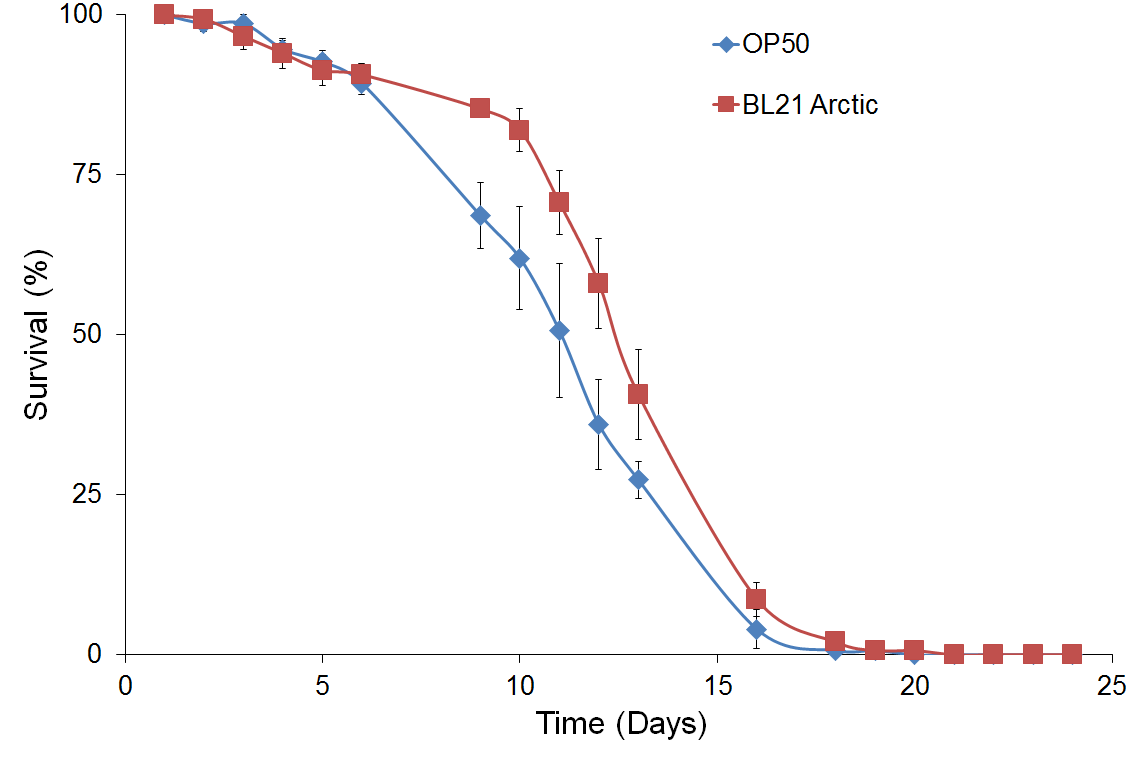
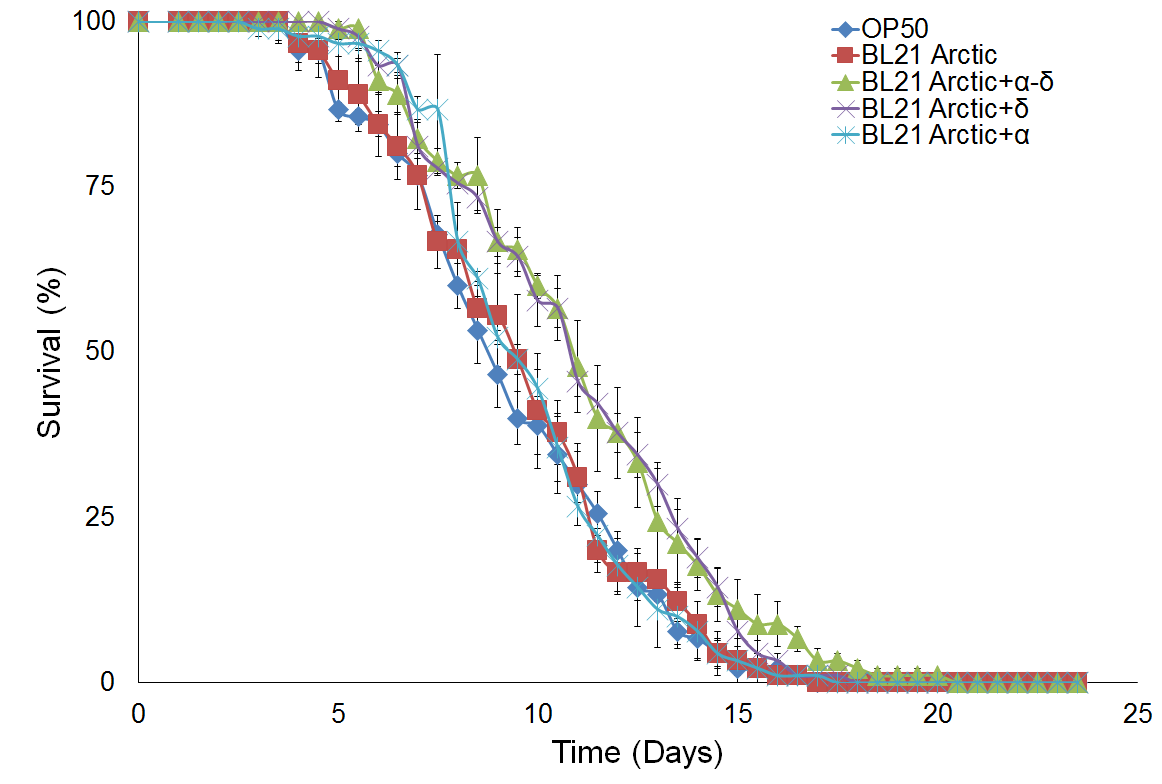
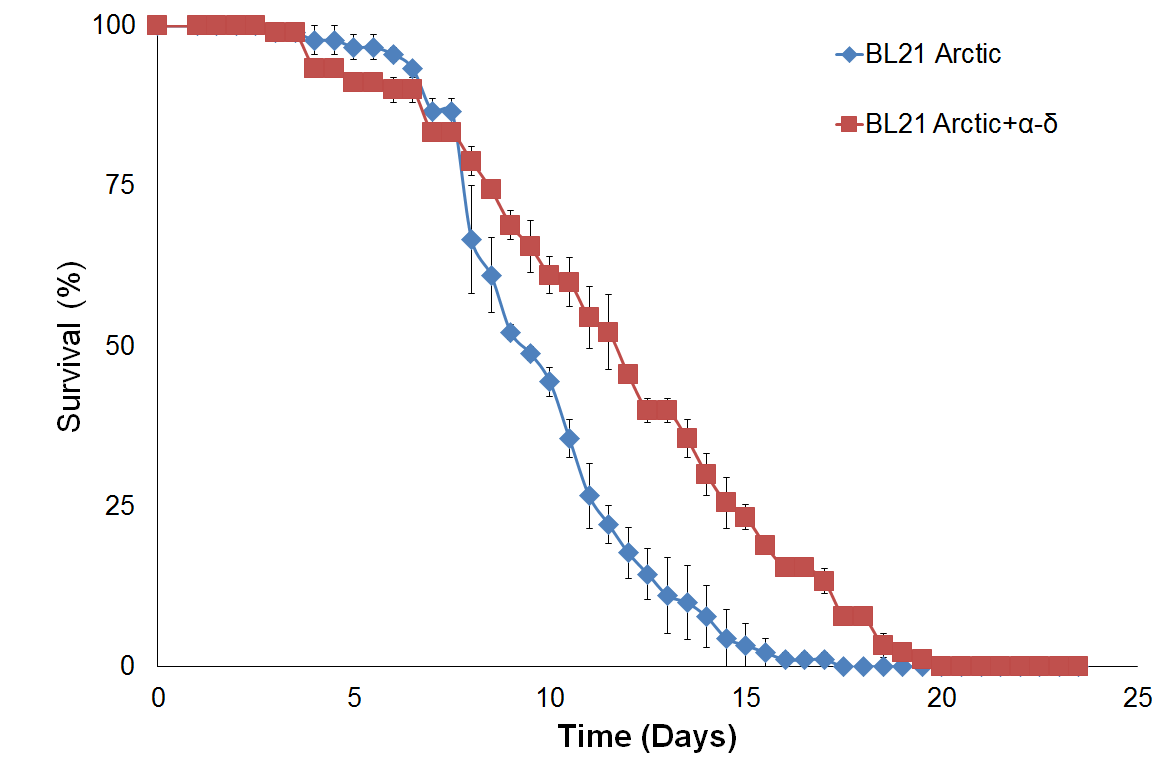
Figure 4. Survival assay with δ protein(s) lawn extends lifespan.
(a) OP50 and BL21 Arctic cells. (b) OP50 and BL21 Arctic cells transformed with capsid construct (α, δ, α-δ) all induced with Rhamnose. Arctic cells expressing α-δ and δ extend lifespan. (c) Survival assay of C. briggsae, BL21 Arctic cells expressing α-δ have increased lifespan.
2.3. Orsay virus capsid proteins affects gut structural and morphological features
Six different features were defined to quantify the intestinal defects observed after infection with OrV proteins as shown in Figure 5. The width of the lumen measured at different points is often smaller. An enlargement, defined as a local widening of the gut, is frequently observed. Spreading as an uneven presence of fluorescence along the digestive system is also observed at a higher frequency. Many undigested contents including integral bacteria inside the lumen are seen. We also observed twisting defined as a twirling of the intestine. Obstructions were also present when the food was in the upper part of the intestine. We assessed those features induced by protein feeding over time. Differences were observed between 4 hours’ post-infection to over at least 1 day after infection (P < 0.05) in all the experiments.
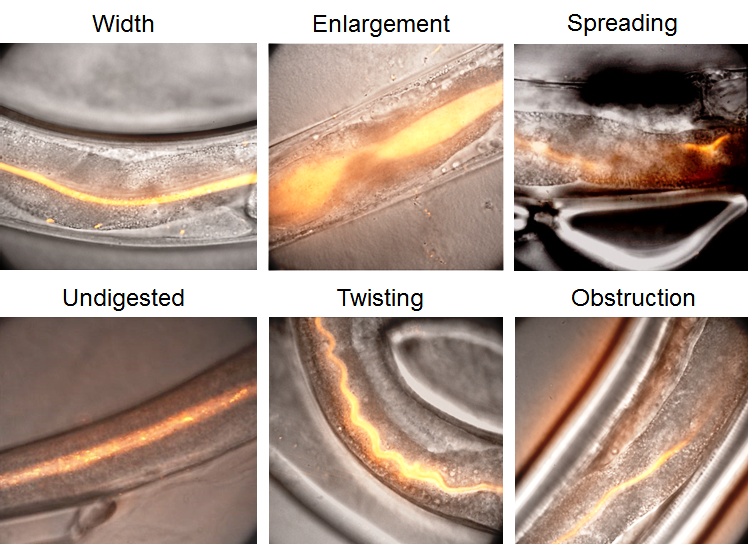
Figure 5. Protein α-δ affects gut morphological features.
Images of the intestinal tract of C. elegans showing the various types of morphologies that were frequently observed. The width of the lumen measured at different points varied. An enlargement, defined as a local widening of the gut. Spreading is an uneven presence of fluorescence along the digestive system. Many undigested contents including integral bacteria inside the lumen can be seen. Twisting is defined as a twirling of the intestine. Occlusion can also be seen when the food is only present in the upper part of the intestine. (n = 36, synchronized population over 5 days).
Phenotypic analysis and scoring of these features are shown in Figure 6 and indicate a narrowing of the intestinal lumen in the presence of the capsid proteins α, δ or α-δ (P < 10-4, ANOVA and Tukey-HSD test). The protein α has a stronger effect on the narrowing of the lumen in comparison to δ or α-δ (P < 10-4, ANOVA and Tukey-HSD test). The local enlargement of the intestine was less seen in the nematodes fed with the viral protein α, δ or α-δ, compared to the control (P < 10-4, = 10-4, = 2x10-3, respectively, Tukey-HSD tests). Even though no statistical differences in the number of twists were observed between the control and α-δ and δ, the nematode α-fed showed a decrease in the numbers of twists (P = 7x10-3, Tukey-HSD tests). The protein α seems to prevent the contraction of the worm intestine. The distribution of the food inside the lumen was also affected by the viral proteins, becoming less uniform with the viral protein. Interestingly, spreading was observed more in nematode fed with α-δ and α (P = 1x10-3, 2x10-3, respectively, Tukey-HSD tests) but not δ. The presence of undigested food at the end of the intestinal tract was only significantly different in the worm fed with the fusion protein α-δ (P < 10-4, Tukey-HSD tests). However, the occlusion by the food does not seem to be responsible for any particular protein as no differences were observed between each condition (all P = 0.5, Kruskal-Wallis test). Taken together, the protein α has a greater effect on the narrowing of the lumen, based on either the food intake or the speed of digestion. The fusion protein α-δ affects mostly an uneven distribution of digested or undigested food along the digestive tract; therefore, it could play a role in the global digestion ability of the nematode. The protein δ is not associated with any particular feature but contributed to the mal digestion of the nematode.
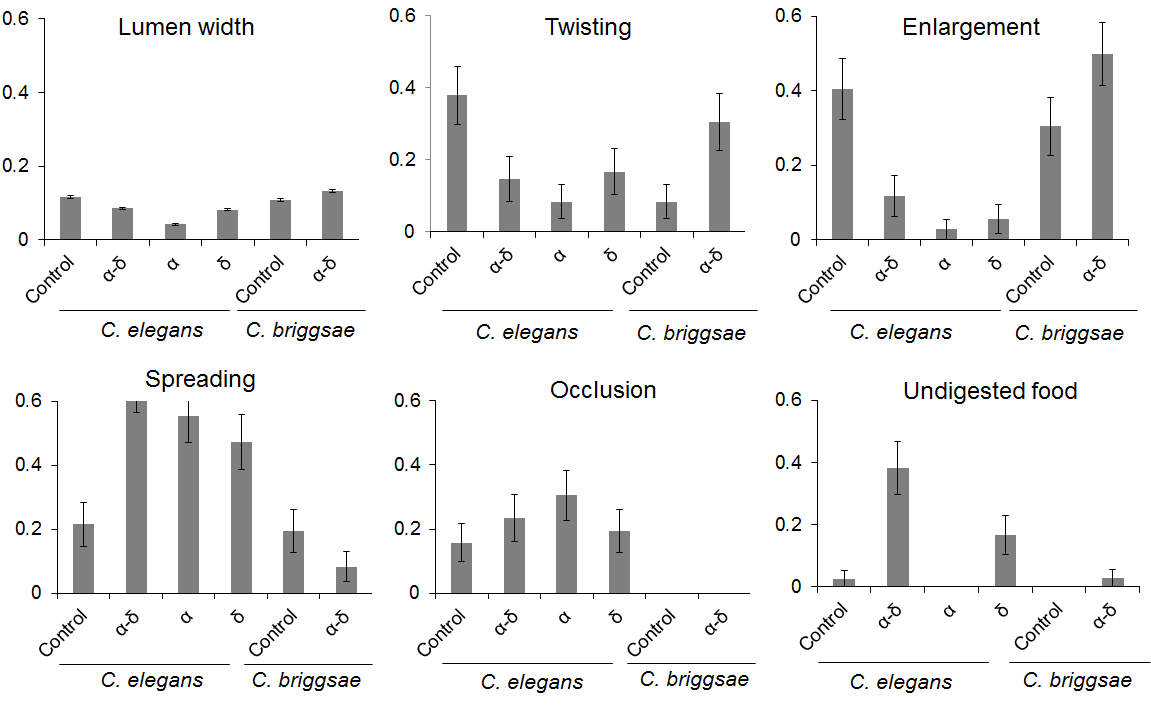
Figure 6. Protein α-δ drives digestive abnormalities in C. elegans.
The control corresponds to BL21 Arctic expressing only the FresnoRFP; the other mentioned the protein fused to the FresnoRFP. In the case of the lumen width, the y-axis corresponds to the lumen intestine width that was normalized against the average total width of the nematode measured. For the other charts, the y-axis represents the fraction of the worm population presenting the feature averaged over replicates. The lumen width is significantly reduced in worm fed with α than with δ or with α-δ. Less twisting was observed in presence of α than in the other conditions. Less enlargements were seen when C. elegans was fed with the viral proteins. More spreading and undigested food was detected in the presence of the fusion protein. The obstruction was not able to differentiate the different groups. For C. briggsae, the fusion protein induced only a larger lumen.
In order to understand better, the digestive abnormalities and the role of proteins, correlations between the identified features were assessed for each protein by plotting the principal components, as shown in Figure 7. In the control case, C. elegans has a larger intestine when able to digest food along with an even distribution of the food along the digestive tract. The nematodes δ fed shows a larger intestine and local enlargement when the food is not digested. The nematodes fed with α have a higher presence of twisting and enlargement. Interestingly, the features enlargement and width were uncorrelated in the presence of α; while these two features were correlated in the control case. In the same way, the features associated mostly with the feeding of α-δ, spreading and undigested food, were uncorrelated when the nematodes were fed with them.
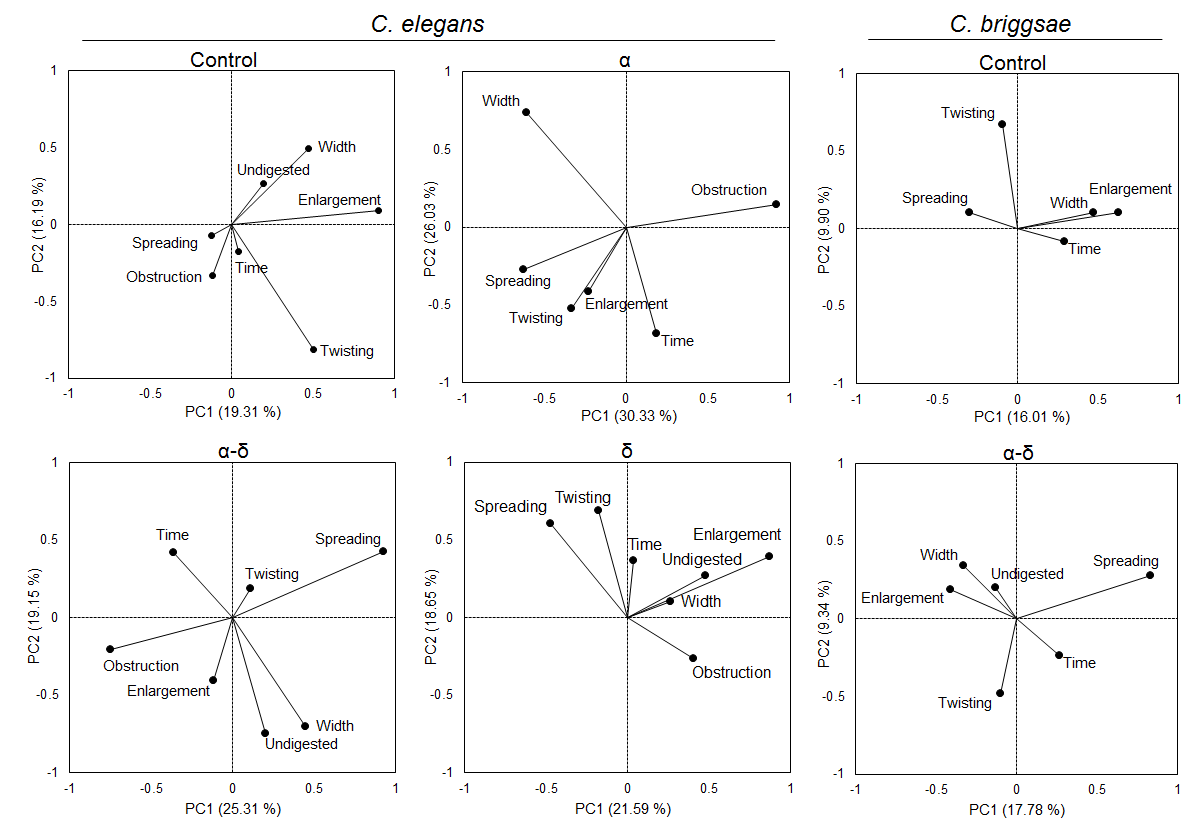
Figure 7. Protein δ correlates gut morphological feature abnormalities.
For C. elegans, in the control, the presence of spreading was negatively correlated with the undigested content and the lumen width correlated with the presence of enlargement. However, in the presence of the protein α, the width and enlargement were uncorrelated while α-δ is mostly corelated for width and undigested but not enlargement features. If some undigested food is accumulated, it will lead to width change and enlargement. On the contrary, α alone or with δ disrupt these corelated features. Uncorrelation was also observed for spreading and undigested contents in the presence of δ and α-δ. For C. briggsae, the width and enlargement stayed correlated in the presence of α-δ. However, spreading became negatively correlated with the presence of enlargement in the presence of the fusion protein.
Taken together, the protein α is responsible for an accumulation of undigested food in the intestine, as observed from the lack of correlation between the two features enlargement and lumen width, therefore, the worms seem to have a lower ability to absorb food. However, the fusion protein α-δ plays a role in the digestion ability, demonstrated by the abnormal presence of food spreading and undigested food along the intestine, two features that appeared together in the control case (Figure 7).
The same studies were performed for C. briggsae, which behaved in the same way as C. elegans, but as opposite responses for twisting, enlargement, spreading and undigested food. In addition, the obstruction feature was not observed (Figure 6). However, the protein α-δ was responsible for a larger intestine and was the only statistical difference observed. Considering the correlation between the features, enlargement was correlated with the larger lumen in both cases. Whereas the presence of spreading was uncorrelated with enlargement in the control case, it was negatively correlated in the worms fed with α-δ.
2.4. Orsay virus proteins promote hyperactive worm behavior
The OrV infection in C. elegans induces a wide range of intestinal symptoms as well as a behavioral change expressed by an increase of motility in OP50 medium infected by OrV, capsids proteins α, δ or α-δ. The difference in behavior between OP50 and BL21 Arctic were notable (data not shown). Previous studies have shown that the bacterial species used to make lawns for feeding, affects C. elegans growth and survival(28). Therefore, different worm behavior strategies could be used by the organism to forage those different types of food. Eight variables measured during the tracking to define the motility parameter values characteristic of this behavior have been plotted in Figure 8.
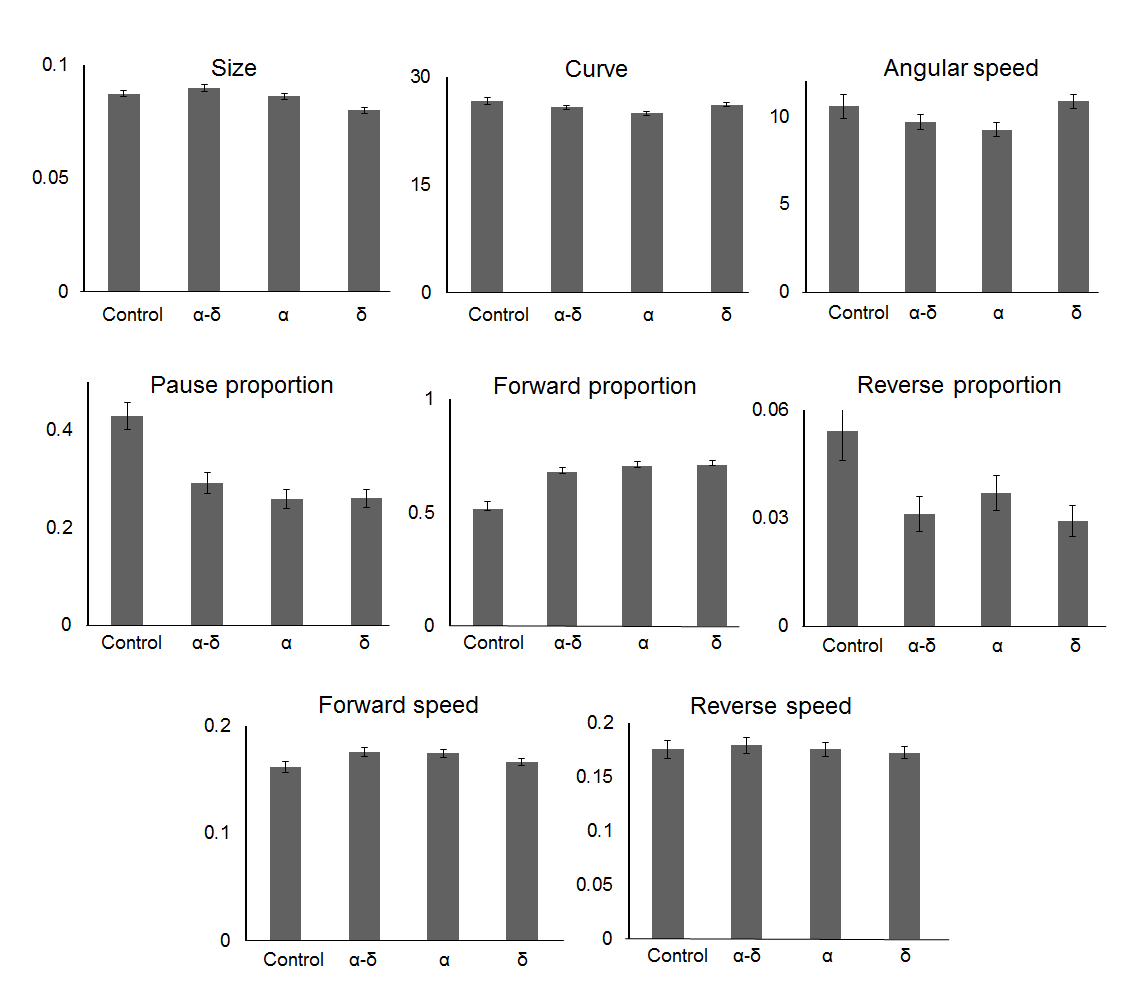
Figure 8. Orsay virus capsid proteins promotes hyperactive behaviour.
In the presence of all the viral proteins, the nematodes present more forward and less reverse movement, moreover they spent less time pausing. The protein δ is involved in development of impediment underlined by a smaller body size, whilst the protein α is responsible for a more linear movement of the worms, characterised by a less curved movement and emphasized by the difference of angular speed between worms α-fed and δ-fed. For y-axis, see material and methods section.
The forward and reverse speed did not differentiate the behavior of the nematodes in the different conditions (P = 0.50 and 0.72 respectively, ANOVA test) (for the control, n = 224, for α, n = 373, for δ, n = 272, for α-δ, n = 401, obtained in three independent experiments). The proportion of worms going forward does differentiate the worms in the presence of α, δ, or α-δ from the control (P < 10-4, ANOVA and Tukey HSD tests). Similar results obtained for the pausing proportion, showed that the worms exposed to the viral proteins spent less time pausing (P < 10-4, ANOVA and Tukey HSD tests). The results were also notable for the proportion of worms in reverse motion, (P = 4x10-4, ANOVA test), yet, only the worms in the presence of δ or α could be differentiated (P = 1x10-3 and 3x10-2, respectively, Tukey HSD tests). Features that are more specific to one protein were also observed. The size of the nematode was affected by the protein α-δ (P < 10-4, ANOVA test and P = 6x10-3, Tukey HSD test) in comparison to the control. This protein was also responsible for less curving movement (P = 3x10-2, ANOVA test and Tukey HSD test) associated with a slower angular speed (P < 10-4, ANOVA test and P = 8x10-4, Tukey HSD test) compared to the control. The presence of protein α-δ was correlated with the curving features of the worms. Proteins α and δ individually were solely correlated with a reduction of reverse proportion.
A habituation assay was also performed where the nematodes presented the same reaction to a succession of taps (data not shown) suggesting that the viral proteins did not have a detectable effect on their response to stress.
2.5. Orsay virus capsid protein δ displayed structural similarity to Human Tryptophan 2,3-dioxygenase
Optimal structural superposition analysis of viral capsid protein δ (PPDB Id: 5W82) with human TDO (PDB Id: 4PW8), human TDO inhibitor complex (PDB Id: 6A4I), human TDO in complex with tryptophan (PDB Id: 5TIA) and human TDO in complex with tryptophan and dioxygen (PDB Id: 5TI9) illustrated that the capsid δ have high degree of structural similarity with the human TDO structure (Figre 9). The structure of protein δ aligned well with TDO in the region closed to ALD motif (Root Mean Square Deviation, RMSD = 2.74 Å for 128 equivalent Cα, 88% coverage) but since the second helix has a hinge following a shorter helix, the orientation of the equivalent helix may be different in context of protein δ than the TDO. The final alignment was refined by determining a core of well-aligned atoms and minimizing the RMSD of this core. Both the RMSD value and the number of well-aligned atoms were considered to measure the quality of the alignment. It is significant that both structures have similar length helix structures on the region covering protein δ structure in TDO.
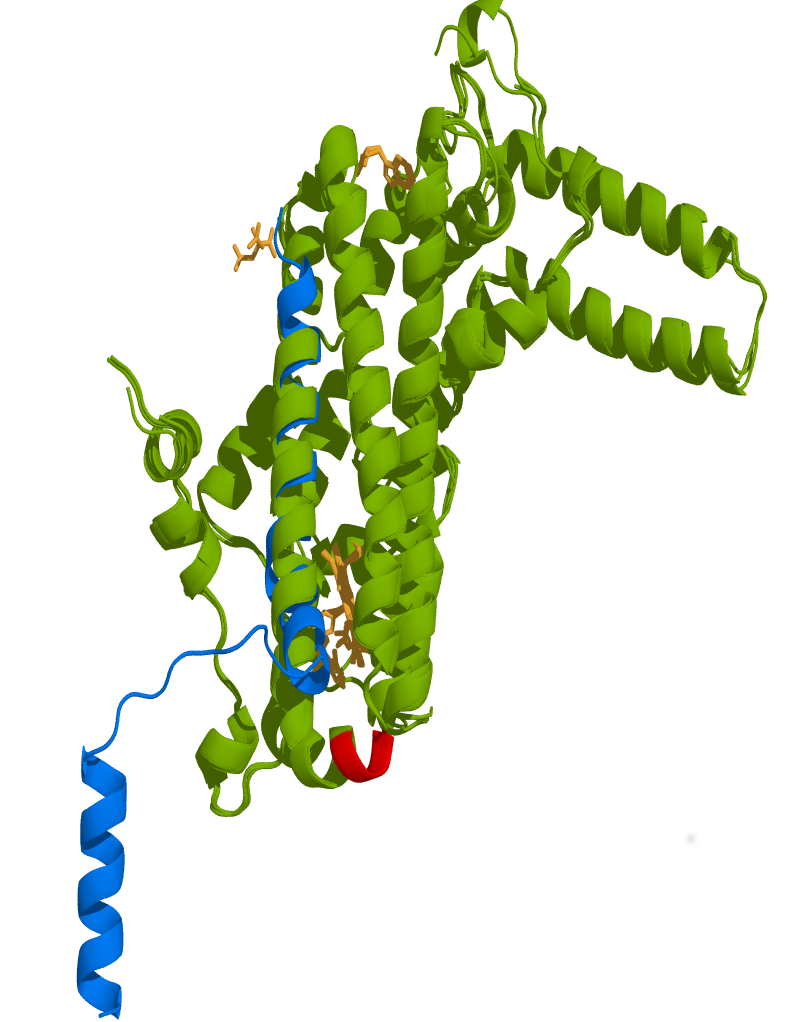
Figure 9. Structural superposition of Orsay virus capsid protein δ to human TDO displays high similarity.
The protein δ presented structural features that are very similar to human TDO suggesting that protein δ could alter TDO activity by acting as a mimetic protein interfering with TDO structure. Green: Human TDO in complex with tryptophan (39-300, PDB Id 5TIA), Human TDO (PDB Id 4PW8), Human TDO inhibitor complex (PDB Id 6A4I), and Human TDO in complex with tryptophan and dioxygen (PDB Id 5TI9). Blue: Viral capsid protein δ (PDB Id 5W82). Red: ALD motif in human TDO essential for its activity. Deletion of this motif (PLD motif in TDO-2) in C. elegans increases mobility and extends lifespan. Orange: Heme active site of human TDO in complex with Tryptophan and dioxygen. The structure of protein δ (12-333) aligned well with TDO (RMSD = 2.74 Å for 128 equivalent Cα, 88% coverage) in the region closed to ALD motif but since the second helix has a hinge following a shorter helix the orientation of the shorter equivalent helix may be different in the context of protein δ than TDO. It is remarkable that both structures have similar length helix structures on the region covering protein δ structure in TDO.
3. Discussion
The α-δ fusion cDNA used for E. coli expression was codon-optimized for this bacterial species. As a result, the viral cDNA sequence is different from the original OrV cDNA and may not be as efficient to elicit the RNAi response. Our result showed that the proteins are more soluble at a lower temperature. Given the strategies used to fuse the GFP downstream the cDNA and the fact that engineered bacterial patches α, δ or α-δ were highly fluorescent, those results strongly support the concept that the capsid proteins were folded(25). Unfolded protein from cloning of cDNA upstream from the GFP-cDNA would not produce folded RFP and as a result would not emit fluorescence. Because the capsid proteins are fused to RFP and are fluorescent, we can conclude that the protein is folded. This protein feeding methodology provided a direct readout of the effect of the capsid proteins to the gut lining of the worm. Moreover, in the study of 1, OrV infection in a rde-1 (+) strain showed no phenotype suggesting that C. elegans controlled the spread of infection by using the RNAi response. Our results are similar to the observations in the rde-1(-) strain and emphasize the strong contribution of capsid proteins to the phenotype observed. Survival curve assays provided additional evidence for the important role of these proteins in C. elegans lifespan, where we showed that protein δ contributed the most to worm’s lifespan extension. The protein α induced a muscular impairment seen by a specific linear movement and a mal digestion. Indeed, if the digestive muscles do not work properly, the contraction of the intestinal cells would be impaired and would create an abnormal and inconstant digestion. Such as, the abnormal distribution of the food is seen through the lack of spreading and affection of the intestinal flexibility of the digesting tracts underlined through the lack of enlargement and twisting (Figure 6). With the protein α-δ, the spreading of food in the intestine was more prevalent and significant presences of undigested food were seen, suggesting a higher effect of the intestinal muscles that would not induce an irregular digestion but a mal-digestion. In the presence of any of the viral protein, the worm moved more forward with less reverse movement and less pausing suggesting a will of the worm to find a more edible food or a better environment even though the speed was not affected. The effects of the capsid proteins were more important in the intestine, which is not surprising as OrV infects intestinal cells. Previous reports mentioned a disorganization of the intestinal cell structure, even though we did not measure them, we could observe some of them and this was underlined by the reduction of the ability of the worm to digest. Moreover, those symptoms were not observed in the case of C. briggsae, a non-specific host to Orsay virus (data not shown), showing the ability of the tested proteins to impact intestinal function in C. elegans. The behavioral hyperactivity observed using Multi-Worm Tracker can only be accounted for by the proteins α, δ and α-δ and that those proteins are the driver of an adaptive behavioral response to food ingestion and locomotion. The timing in the Multi-Worm Tracker experiment that showed change of behavior (in less than a day) are consistent with the stimulation of sensory neuron by serotonin(15,20), suggesting that a serotonin response may be involved in this change of behavior induced by capsid proteins that is further under investigation in our lab.
Different hypotheses can also be proposed to explain our observations. One could be that α, δ and α-δ proteins affect serotonin (5-HT) secretion of the cells that can detect feeding cues (roaming and dwelling) in the gut, and consequently change their adaptive behavioral response to food ingestion(29). In this study, we observed important morphological changes in the gut of the worm fed by protein α-δ. As a result, lining gut cells that integrate internal state by secreting 5-HT could have altered the ability to target downstream target neurons that drive the adaptive behavioral responses to food ingestion(30). Serotonin-sensitive neurons, in response to food ingestion, can be modulated by internal state(31), that would justify an interpretation of the behavioral observations in our study that we are anticipating at large from our continuing learning. Moreover, protein structural superposition analysis of Orsay virus capsid protein δ with human TDO displayed similar structural features suggesting that viral protein delta may interfere with worms TDO activity by acting as a mimetic protein interfering with the ALD motif in the structure of TDO essential for its function. This dysregulation of TDO in the Kynurenine pathway could favor serotonin production and/or accumulation of kynurenic acids resulting in brain excitability and lifespan extension(21,22).
A second possibility could be to conclude that α-δ remodels C. elegans gut microbiota and modulates 5-HT secretion due to the release by microbiota of 5-HT agonists such as tryptamine that would interfere with the 5-HT response that may affect downstream target sensory neurons. Our system cannot evaluate this possibility since our experimental conditions were likely a germ-free environment. However, a recent study on wild C. elegans microbiota revealed that OrV can shift microbiota bacterial composition(32). We carried out a careful analysis of the type of bacteria that were shifted in this study and observed that some of those species were Bacteroides (Data not shown). Because Bacteroides have been shown to provide 5-HT agonist such as tryptamine that may be used by 5-HT downstream sensory neuron <(33, 34), α-δ could indirectly affect downstream sensory neurons to modulate behavioral hyperactivity observed in this study.
In conclusion, given the increasing interest to identify key features of the gut-brain axis in the worm, our study presented convincing and novel finding showing that viral proteins can affect behavior and lifespan through gut interactions and play an important role in virus pathogenicity as virulent factors. In a broader sense, the method developed in this work will allow studies of the effect of any protein on a worm gut in a spatio-temporal manner. It could also be used as a tool to engineer bacterial strains that can express any specific set of proteins to better understand how microbiota proteins can interact with worm behavior. The application of this approach in the future may provide novel discoveries in the field of host-microbiota interaction in the C. elegans model organism also of relevance to human.
4. Materials and Methods
4.1. Nematode strains and maintenance
The reference wild-type N2 strain of C. elegans (Bristol, UK) and AF16 strain of C. briggsae wild isolate (Ahmedabad, India) were obtained from the Caenorhabditis Genetics Centre (University of Minnesota, Minneapolis, MN, USA). Laboratory strains were maintained via standard laboratory techniques on Nematode Growth Media (NGM) plates seeded with E. coli OP50, the normal laboratory diet. To obtain comparative results all experiments were carried out using synchronized worm populations.
4.2. Gene synthesis and codon optimization of Orsay virus capsid cDNA
The OrV nucleotide sequence isolated from the wild strain nematode JU1580 (GenBank Accession Number: NC_028097) was obtained from NCBI. The c-DNA α-δ fusion protein was synthesized from this sequence and codon-optimized by DNA2.0 company now ATUM (Newark, CA, USA). The resulting modified α-δ fusion cDNA sequence obtained by codon optimization was 77% similar to the original sequence at the nucleotide level (Sequence available upon request). The α-δ cDNA was used as a template to further sub-clone α and δ in different expression vectors for protein expression.
4.3. Capsid recombinant proteins – Sub-cloning and expression in E. coli
The optimized cDNA α-δ fusion was first integrated into the protein expression vector pD441-SR (DNA 2.0), which is a high copy vector characterized by kanamycin resistance and an IPTG-inducible T5 promoter to ensure a high level of protein expression. The plasmid was then transformed into BL21 Arctic cells, which allows for the tuning of the level of protein expression by decreasing the temperature of expression to find the optimal conditions that would give the highest yield of soluble protein. Effectively, this E. coli strain was genetically rewired in such a way that protein could be expressed and correctly folded at very low temperature (4-12 ºC). This was feasible due to genome insertion of a thermostable chaperonin that can refold protein at this extreme temperature. It provided us with a wider range of temperature to explore for solubility studies. The solubility of the protein α / δ was measured by comparing the level of protein expression in the whole fraction, in the supernatant, and in the cell pellet at different temperatures 11.5 ºC, 20 ºC and 37 ºC. Since α-δ fusion protein was overproduced, the α- and δ- cDNA was sub-cloned into the vector pD861-SR (DNA 2.0) for protein expression optimization. This vector contains a promotor inducible by Rhamnose from which the amount of protein produced could be controlled by varying the concentration of Rhamnose during protein induction and eventually limiting the production of the inclusion body by slowing down the process. α and δ cDNA were sub-cloned into pBAD vector using the Sap-I restriction site in the multiple cloning site cassette by PCR amplification of the template α-δ cDNA optimized sequence using a specific set of primers Sap I-AF, Sap I-AR, Sap I-DF and Sap I-DR (Table 1).
Table 1. Details of primers used in this studya.
|
Product
|
Gene
|
Primer name
|
Primer sequence 5-3´
|
|
Amplification
|
α
|
Sap 1-AF (Forward)
|
TACACGTACTTAGTCGCTGAAGCTCTTCTATGAACAAAAACAAC
|
|
SAP 1-AR (Reverse)
|
TAGGTACGAACTCGATTGACGGCTCTTCTACCGATATTGGAGCTCGGCAGGCTGTA
|
|
δ
|
SAP 1-DF
(Forward)
|
TACACGTACTTAGTCGCTGAAGCTCTTCTATGGTGAAACTGGGC
|
|
SAP 1-DR
(Reverse)
|
TAGGTACGAACTCGATTGACGGCTCTTCTACCTTATTAGTCATTACGCGTCGCCAG
|
|
Amplification & Stitching
|
FresnoRFP
|
Sap1-S
(Forward)
|
TACACGTACTTAGTCGCTGAAGCTCTTCTATG
|
|
FresnoRFP-F-AD-R
(Reverse)
|
CTTATGGGTGTTGTTTTTGTTCATTTTGTACAGTTCGTCCAT
|
|
α-δ
|
FresnoRFP-F-AD-F
(Forward)
|
ATGGACGAACTGTACAAAATGAACAAAAACAACACCCATAAG
|
|
Sap1-E-AD
(Reverse)
|
AGGTACGAACTCGATTGACGGCTCTTCTACCTTAGTCATTACGCGTCGCCAG
|
|
α
|
FresnoRFP-F-AD-F
(Forward)
|
ATGGACGAACTGTACAAAATGAACAAAAACAACACCCATAAG
|
|
Sap1-A
(Reverse)
|
TAGGTACGAACTCGATTGACGGCTCTTCTACCCTACTAATTGGAGCTCGGCAGGCTGTA
|
|
δ
|
Fresno-F-D-F
(Forward)
|
ATGGACGAACTGTACAAAATGCCGAGCGAAGATTAC
|
|
Sap1-E-AD
(Reverse)
|
AGGTACGAACTCGATTGACGGCTCTTCTACCTTAGTCATTACGCGTCGCCAG
|
|
FresnoRFP-α-δ and FresnoRFP-δ
|
Sap1-S
(Forward)
|
TACACGTACTTAGTCGCTGAAGCTCTTCTATG
|
|
Sap1-E-AD
(Reverse)
|
AGGTACGAACTCGATTGACGGCTCTTCTACCTTAGTCATTACGCGTCGCCAG
|
|
FresnoRFP-α
|
Sap1-S
(Forward)
|
TACACGTACTTAGTCGCTGAAGCTCTTCTATG
|
|
Sap1-A
(Reverse)
|
TAGGTACGAACTCGATTGACGGCTCTTCTACCCTACTAATTGGAGCTCGGCAGGCTGTA
|
aSap-I was used as a restriction site wherever applicable.
4.4. Capsid proteins fused to GFP protein variants - Cloning and expression in E. coli
To determine which GFP (Green Fluorescent Protein) variants would give us the most stable and strongest fluorescent signal when fused to OrV capsid proteins, we made different constructs by stitching α-δ cDNA to plasmid protein expression vector containing GFP, YFP (Yellow Fluorescent Protein) and FresnoRFP (Red Fluorescent Protein). Our results suggested strongly that FresnoRFP would be the best candidate. We could easily see the pink coloration in solid and liquid media retained in the bacterial pellet when the protein was expressed. Furthermore, confocal microscopy revealed that FresnoRFP was giving the strongest, stable and durable fluorescence emission in the gut of the worms.
Different constructs with the expression vector pD861-SR inducible by Rhamnose were made by fusing the cDNA region of α and δ to FresnoRFP. In our construct design, the gene fragment of interest was cloned 5ʹ- upstream the FresnoRFP sequence to assess proper protein folding according to previous studies (25, 35). In this orientation, if the insert cDNA does not fold properly it is unlikely that the GFP would fold at all and as a result, no fluorescence would be obtained. A negative control was also made using a plasmid vector expressing FresnoRFP only. The resulting constructs were transformed into BL21 Arctic cells. From the construct with FresnoRFP, the expression of the protein was assessed by looking at the fluorescent signal.
Amplifications of the FresnoRFP and α-δ, α, and δ proteins were performed by PCR amplification as previously using specific primers that containing a Sap-I (forward) restriction-cloning site (Table 1). The PCR products were purified using a QIAquick PCR purification kit (Qiagen) following the manufacturer’s protocol. The correct DNA insert was confirmed by sequencing (Genewiz). The expression and solubility of the protein was confirmed by SDS-PAGE of total, cell pellet, supernatant fraction.
4.5. Worm survival assay from bacterial lawn expressing capsid proteins
To assess the toxicity of worms fed with different bacterial lawn expressing the capsid proteins, survival assays were performed from synchronized populations(36). For each experimental condition, 30 synchronized worms were picked and transferred to NGM plates seeded with E. coli OP50 or E. coli BL21 Arctic bacterial cells (previously transformed with capsid proteins expression vectors) and incubated at 25 ºC. To maintain plasmid selection in the lawn and express the protein, bacteria transformed with the vector pD861-SR, were grown on a modified NGM media containing L-Rhamnose monohydrate (1 mM) and Kanamycin monosulfate (25 µg ml-1). In the presence of the vector pD444-SR, the NGM media was modified by adding IPTG (1 mM) and Ampicillin (100 µg ml-1). The cultures were maintained and scored for survival. Immobile worms or worms not responding to gentle agitation were considered dead. Worms burst or in the plastic lid were deemed censored. Survival of the populations was assessed until no worms remained alive and the experiment was done in triplicate. The results were analyzed by Kaplan-Meier analysis using the feature “survival curve analysis” from GraphPad Prism 4.0(37). The curves were drawn using R program in Rstudio(38).
4.6. Capsid proteins localization in C. elegans – Phenotype scoring by confocal fluorescence microscopy
To determine the localization of the capsid proteins:FresnoRFP fusions in the nematode and their contribution to the intestinal abnormalities phenotype, worms fed with individual proteins were observed under differential interference contrast microscopy (DIC) and fluorescence microscopy using the 40x objective. Six adults, worms were observed each day for a week and different variables were derived from preliminary observations, (i) the width of the fluorescence in the lumen corresponds to the lumen intestine width that was normalized against the average total width of the nematode measured at different points along the nematode body, (ii) the presence or absence of lumen enlargement described as a suddenly larger lumen in one particular point of the lumen, (iii) twisting of the gut lining characterized by several twists in the same portion of the lumen, (iv) the presence of undigested food was set when the bacteria were ungraded even in the terminal part of the intestine. Sometimes, bacteria were still moving underlying the fact that they were not digested, (v) the spreading of fluorescence seen by an alternation of presence and absence of fluorescence, and (vi) obstruction of the gut lining when the fluorescence was only present in the upper part of the intestine but neither in the middle nor in the terminal part. The intestinal features were then counted and were analyzed by Kruskal-Wallis and Tukey tests using the Excel plug-in RealStats(39).
Principal Component Analysis (PCA) was performed to correlate and compare worm’s phenotypic features before and after capsid proteins feeding. Firstly, the values of each intestinal feature were standardized and then the PCAs were drawn with these values using package FactoMineR from R studio(38).
4.7. Behavioral tracking after capsid proteins feeding
In many instances, we observed a change of worm locomotion reporting a hyperactive change of behavior when incubated with lawn contaminated with a viral extract from JU1580 as well as worms expressing different capsid proteins not fused to FresnoRFP compared to uninfected worms. Those observations led us to quantify the phenotype more precisely. Worm fed with those lawns were monitored by a MWT system that can analyze and quantify the basic movement responsible for this behavior. Age-synchronized animal populations were reared at 20 °C and behaviorally tracked on the rearing plates at 96 hours after age synchronization. Data collection of worm behavior and movement were made in triplicates using 3 different instruments. Image acquisition and data analyzes were performed using the Multi-Worm Tracker software: version 1.2.0.2, http://sourceforge.net/projects/mwt(40). Plates were tracked for 600 seconds and spontaneous activities of the worms were recorded. Offline analysis with Choreography software: version 1.3.0_r1035(31) used “--shadowless”, “--minimum-time 20”, and “--minimum-move-body 2” filters. Custom MATLAB® (The MathWorks, Natick, MA; http://wormsense.stanford.edu) scripts organized and summarized Choreography output files. In the MATLAB® analysis, data were binned into 60, 10-second intervals, with each data point representing the collective mean of all tested animals (n ~ 300) during the interval. The proportion of locomotion (forward, backward, and pausing), speed of locomotion (forward and backward, mm/s), body size in area (mm2), angular speed (rad/s), and curve (degree) were used to quantify the effects of treatments.
4.8. Capsid protein structural analysis
The protein 3D-coordinates of Orsay virus capsid protein δ and human TDO were retrieved from the Protein Data Bank (PDB) with the following identifiers (Id): Viral capsid protein δ: PDB Id 5W82, Human TDO: PDB Id 4PW8, Human TDO inhibitor complex: PDB Id 6A4I, Human TDO in complex with tryptophan: PDB Id 5TIA and Human TDO in complex with tryptophan and dioxygen: PDB Id 5TI9. Structural comparison by superposition was performed using ALIGN(41) and SUPER algorithms implemented in PyMOL(42). The RMSD value of the atomic coordinates was determined and considered along with the number of well-aligned atoms as criteria for selecting the best superposition(43).
4.9. Statistics
For survival curve comparison the log-rank test method that looks for trends was used in GraphPad Prism 4.0(37). This method reports a chi-square value that is being used to compute a P value to test the null hypothesis. A low P value indicates that there is a significant trend. The P value tests the null hypothesis that the survival curves are identical in the overall populations. In other words, the null hypothesis is that the treatment did not change survival. A one-way Anova with Tukey’s HSD post-hoc tests were used to determine the significance of treatment effects. The collective means of the last 60 seconds (6 data points) were grouped and compared.
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Acknowledgements: This research work was financially supported by Natural Sciences and Engineering Research Council of Canada through NSERC Discovery Grant (R611657) to Dr. Frederic Pio. The authors thank Dr. Catharine Rankin, Professor in the Department of Psychology at the University of British Columbia and her laboratory staff for facilitating the Multi-Worm Tracker experiments, as well as to the Caenorhabditis Genetics Center (CGC) for strains.
References
-
Felix, M.A, et al. Natural and experimental infection of Caenorhabditis nematodes by novel viruses related to nodaviruses. PLoS Biol 9,e1000586 (2011). https://doi.org/10.1371/journal.pbio.1000586 PMid:21283608 PMCid:PMC3026760
-
Jiang, H. et al. Orsay virus utilizes ribosomal frameshifting to express a novel protein that is incorporated into virions. Virology 450-451, 213-21 (2014a). https://doi.org/10.1016/j.virol.2013.12.016 PMid:24503084 PMCid:PMC3969245
-
Fan, Y. et al. Structure of a pentameric virion-associated fiber with a potential role in Orsay virus entry to host cells. PLoS Pathog 13(2), e1006231 (2017). https://doi.org/10.1371/journal.ppat.1006231 PMid:28241071 PMCid:PMC5344674
-
Jiang, H., Franz, C.J. & Wang, D. Engineering recombinant Orsay virus directly in the metazoan host Caenorhabditis elegans. J Virol 88, 11774-81 (2014b). https://doi.org/10.1128/JVI.01630-14 PMid:25078701 PMCid:PMC4178717
-
Ni, P. & Kao, C.C. Non-encapsidation activities of the capsid proteins of positive-strand RNA viruses. Virology. 446(1-2), 123-32 (2013). https://doi.org/10.1016/j.virol.2013.07.023 PMID: 24074574; PMCID: PMC3818703.
-
Weber, P.H. & Bujarski, J.J. Multiple functions of capsid proteins in (+) stranded RNA viruses during plant-virus interactions. Virus Res. 196, 140–149 (2015). https://doi.org/10.1016/j.virusres.2014.11.014 PMID: 25445337.
-
Bol, J.F. Role of capsid proteins. Methods Mol Biol 451, 21-31 (2008).
https://doi.org/10.1007/978-1-59745-102-4_2 PMid:18370245
-
Ding, S.W. RNA-based antiviral immunity. Nat Rev Immunol 10, 632-644 (2010). https://doi.org/10.1038/nri2824
PMid:20706278
-
West, C. & Silverman, N. Drosophilosophical: re-thinking adaptive immunity in the fly. Cell 169, 188-190 (2017).
https://doi.org/10.1016/j.cell.2017.03.032 PMid:28388404
-
Ashe, A. et al. A deletion polymorphism in the Caenorhabditis elegans RIG-I homolog disables viral RNA dicing and antiviral immunity. Elife 2, e00994 (2013). https://doi.org/10.7554/eLife.00994 PMid:24137537 PMCid:PMC3793227
-
Sarkies, P. et al. Competition between virus-derived and endogenous small RNAs regulates gene expression in Caenorhabditis elegans. Genome Res 23, 1258-1270 (2013). https://doi.org/10.1101/gr.153296.112 PMid:23811144 PMCid:PMC3730100
-
Ashe, A., Sarkies, P., Le Pen, J., Tanguy, M. & Miska, EA. Antiviral RNA interference against Orsay virus is neither systemic nor transgenerational in Caenorhabditis elegans. J Virol 89, 12035-46 (2015). https://doi.org/10.1128/JVI.03664-14
PMid:26401037 PMCid:PMC4645334
-
Bakowski, M.A., Desjardins, C.J. & Smelkinson, M.G. Ubiquitin-mediated response to microsporidia and virus infection in C. elegans. PLoS Pathog 10(6), e1004200 (2014). https://doi.org/10.1371/journal.ppat.1004200 PMid:24945527 PMCid:PMC4063957
-
Schier, A.F. Should I stay or should I go: neuromodulators of behavioral states. Cell 154, 955-956 (2013).
https://doi.org/10.1016/j.cell.2013.08.017 PMid:23993087
-
Flavell, S.W. et al. Serotonin and the neuropeptide PDF initiate and extend opposing behavioral states in C. elegans. Cell 154, 1023-1035 (2013). https://doi.org/10.1016/j.cell.2013.08.001 PMid:23972393 PMCid:PMC3942133
-
Arous, B.J., Laffont, S. & Chatenay, D. Molecular and sensory basis of a food related two-state behavior in C. elegans. PLoS One 4, 7584 (2009). https://doi.org/10.1371/journal.pone.0007584 PMid:19851507 PMCid:PMC2762077
-
Shtonda, B.B. & Avery, L. Dietary choice behavior in Caenorhabditis elegans. J Exp Biol 209, 89-102 (2006).
https://doi.org/10.1242/jeb.01955 PMid:16354781 PMCid:PMC1352325
-
Albertson, D.G. & Thomson, J.N. The pharynx of Caenorhabditis elegans. Phil Trans R Soc Lond B Biol Sci 275, 299-325 (1976). https://doi.org/10.1098/rstb.1976.0085 PMid:8805
-
White, J.G. et al. Brenner, The structure of the nervous system of the nematode Caenorhabditis elegans. Philos Trans R Soc Lond B Biol Sci 314, 1-340 (1986). https://doi.org/10.1098/rstb.1986.0056 PMid:22462104
-
Dalliere, N. et al. Caenorhabditis elegans feeding behaviors Oxford Research Encyclopedia of Neuroscience. (2017).
https://doi.org/10.1093/acrefore/9780190264086.013.190 PMCid:PMC5698734
-
Lemieux, GA. et al. Kynurenic acid is a nutritional cue that enables behavioral plasticity. Cell 160, 119-131 (2015).
https://doi.org/10.1016/j.cell.2014.12.028 PMid:25594177 PMCid:PMC4334586
-
Michels, H. et al. Identification of an evolutionary conserved structural loop that is required for the enzymatic and biological function of tryptophan 2,3-dioxygenase. Sci Rep 6, 39199 (2016). https://doi.org/10.1038/srep39199 PMid:27995966 PMCid:PMC5171515
-
Ball, L.A. & Johnson, KJ. Reverse genetics of nodaviruses. Adv. Virus Res 53, 229-244 (1999).
https://doi.org/10.1016/S0065-3527(08)60350-4
-
Balla, K.M. & Troemel, E.R. Caenorhabditis elegans as a model for intracellular pathogen infection. Cell Microbiol 15, 1313-1322 (2013). https://doi.org/10.1111/cmi.12152 PMid:23617769 PMCid:PMC3894601
-
Waldo, G.S. et al. Rapid protein-folding assay using green fluorescent protein. Nat Biotechnol 17,691-695 (1999).
https://doi.org/10.1038/10904 PMid:10404163
-
Pang, S. & Curran, S.P. Adaptive capacity to bacterial diet modulates aging in C. elegans. Cell Metab 19, 221-31 (2014).
https://doi.org/10.1016/j.cmet.2013.12.005 PMid:24440036 PMCid:PMC3979424
-
Kumar, S. et al. Lifespan extension in C. elegans caused by bacterial colonization of the intestine and subsequent activation of an innate immune response. Dev Cell 49(1), 100-117 e6 (2019) https://doi.org/10.1016/j.devcel.2019.03.010 PMID: 30965033; PMCID: PMC6946027
-
So, S. et al. Control of lifespan by food bacteria, nutrient limitation and pathogenicity of food in C. elegans. Mech Ageing Dev 132, 210-212 (2011). https://doi.org/10.1016/j.mad.2011.02.005 PMid:21354440
-
Rhoades, J.L. et al. ASICs mediate food responses in an enteric serotonergic neuron that controls foraging behaviors. Cell 176, 85-97 (2019). https://doi.org/10.1016/j.cell.2018.11.023 PMid:30580965 PMCid:PMC6526957
-
Bellono, N.W. et al. Enterochromaffin cells are gut chemosensors that couple to sensory neural pathways. Cell 170, 185-198 (2017). https://doi.org/10.1016/j.cell.2017.05.034 PMid:28648659 PMCid:PMC5839326
-
Song, B.M. & Avery, L. Serotonin activates overall feeding by activating two separate neural pathways in Caenorhabditis elegans. J Neurosci 32, 1920-1931 (2012). https://doi.org/10.1523/JNEUROSCI.2064-11.2012 PMid:22323705 PMCid:PMC3463504
-
Guo, Y. et al. The shift of the intestinal microbiome in the innate immunity-deficient mutant rde-1 strain of C. elegans upon Orsay virus infection. Front Microbiol 8, 933 (2017). https://doi.org/10.3389/fmicb.2017.00933 PMid:28611740 PMCid:PMC5446984
-
Yano, J.M. et al. Indigenous bacteria from the gut microbiota regulate host serotonin biosynthesis. Cell 161, 264-76 (2015).
https://doi.org/10.1016/j.cell.2015.02.047 PMid:25860609 PMCid:PMC4393509
-
Zhang, F. et al. Caenorhabditis elegans as a model for microbiome research. Front Microbiol 8, 485 (2017). https://doi.org/10.3389/fmicb.2017.00485 PMid:28386252 PMCid:PMC5362939
-
Lau, D. et al. Design and selection of IFI16-PAAD mutants with improved dsDNA destabilization properties. J Proteomics Bioinform 9, 255-263 (2016). doi: 10.4172/jpb.1000414
-
Lionaki, E & Tavernarakis, N. Assessing aging and senescent decline in Caenorhabditis elegans: cohort survival analysis. Methods Mol Biol 965, 473-84 (2013). https://doi.org/10.1007/978-1-62703-239-1_31 PMid:23296678
-
Miller, J.R. GraphPad Prism Version 4.0 Step-by-Step Examples, GraphPad Software Inc., San Diego CA; (2003).
-
RStudio Team, RStudio: Integrated development for R. RStudio, Inc., Boston MA, http://www.rstudio.com/; (2015).
-
Zaiontz, C. Real statistics using excel. http://real-statistics.com/other-key-distribution/gamma-function, (2016).
-
Swierczek, N.A. et al. High-throughput behavioral analysis in C. elegans. Nat Methods 8, 592-598 (2011).
https://doi.org/10.1038/nmeth.1625 PMid:21642964 PMCid:PMC3128206
-
Cohen, G.E. ALIGN: a program to superimpose protein coordinates, accounting for insertions and deletions. J Appl Crystallogr 30, 1160-1161 (1997). https://doi.org/10.1107/S0021889897006729
-
DeLano, W.L. The Pymol molecular graphics system. DeLano Scientific, San Carlos, CA, USA. http://www.pymol.org (2002).
-
Mishra, P. et al. Meta-analysis suggests evidence of novel stress-related pathway components in Orsay virus - Caenorhabditis elegans viral model. Sci Rep 9,4399 (2019). https://doi.org/10.1038/s41598-019-40762-9 PMid:30867481 PMCid:PMC6416287
|